1BZT
 
 | |
1BZU
 
 | |
1BZV
 
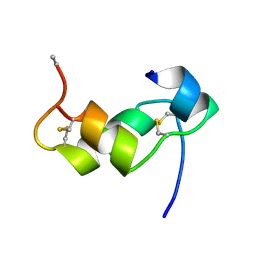 | | [D-ALAB26]-DES(B27-B30)-INSULIN-B26-AMIDE A SUPERPOTENT SINGLE-REPLACEMENT INSULIN ANALOGUE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INSULIN | | Authors: | Kurapkat, G, Siedentopf, M, Gattner, H.G, Hagelstein, M, Brandenburg, D, Grotzinger, J, Wollmer, A. | | Deposit date: | 1998-11-04 | | Release date: | 1999-05-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a superpotent B-chain-shortened single-replacement insulin analogue.
Protein Sci., 8, 1999
|
|
1BZW
 
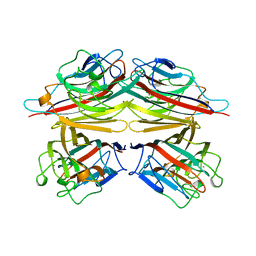 | | PEANUT LECTIN COMPLEXED WITH C-LACTOSE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Surolia, A, Vijayan, M, Lim, S, Kishi, Y. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preferred Conformation of C-Lactose at the Free and Peanut Lectin Bound States
J.Am.Chem.Soc., 120, 1998
|
|
1BZX
 
 | | THE CRYSTAL STRUCTURE OF ANIONIC SALMON TRYPSIN IN COMPLEX WITH BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | CALCIUM ION, PROTEIN (BOVINE PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Leiros, I, Berglund, G.I, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of anionic salmon trypsin in complex with bovine pancreatic trypsin inhibitor.
Eur.J.Biochem., 256, 1998
|
|
1BZY
 
 | | HUMAN HGPRTASE WITH TRANSITION STATE INHIBITOR | | Descriptor: | HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION, PHOSPHORIC ACID MONO-[5-(2-AMINO-4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-3,4-DIHYDROXY-PYRROLIDIN-2-YLMETHYL] ESTER, ... | | Authors: | Shi, W, Li, C, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 1998-11-05 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of human hypoxanthine-guanine phosphoribosyltransferase in complex with a transition-state analog inhibitor.
Nat.Struct.Biol., 6, 1999
|
|
1BZZ
 
 | | HEMOGLOBIN (ALPHA V1M) MUTANT | | Descriptor: | PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Arnone, A. | | Deposit date: | 1998-11-04 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and functional properties of human hemoglobins reassembled after synthesis in Escherichia coli.
Biochemistry, 38, 1999
|
|
1C01
 
 | | SOLUTION STRUCTURE OF MIAMP1, A PLANT ANTIMICROBIAL PROTEIN | | Descriptor: | ANTIMICROBIAL PEPTIDE 1 | | Authors: | McManus, A.M, Nielsen, K.J, Marcus, J.P, Harrison, S.J, Green, J.L, Manners, J.M, Craik, D.J. | | Deposit date: | 1999-07-13 | | Release date: | 2000-07-19 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | MiAMP1, a novel protein from Macadamia integrifolia adopts a Greek key beta-barrel fold unique amongst plant antimicrobial proteins.
J.Mol.Biol., 293, 1999
|
|
1C02
 
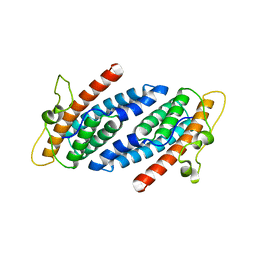 | | CRYSTAL STRUCTURE OF YEAST YPD1P | | Descriptor: | PHOSPHOTRANSFERASE YPD1P | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
1C03
 
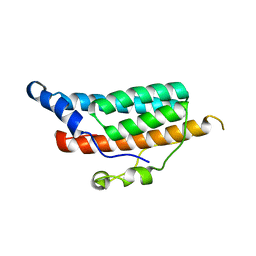 | | CRYSTAL STRUCTURE OF YPD1P (TRICLINIC FORM) | | Descriptor: | HYPOTHETICAL PROTEIN YDL235C | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
1C04
 
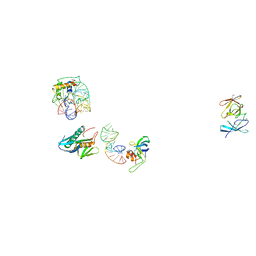 | | IDENTIFICATION OF KNOWN PROTEIN AND RNA STRUCTURES IN A 5 A MAP OF THE LARGE RIBOSOMAL SUBUNIT FROM HALOARCULA MARISMORTUI | | Descriptor: | 23S RRNA FRAGMENT, RIBOSOMAL PROTEIN L11, RIBOSOMAL PROTEIN L14, ... | | Authors: | Ban, N, Nissen, P, Capel, M, Moore, P.B, Steitz, T.A. | | Deposit date: | 1999-07-14 | | Release date: | 1999-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Placement of protein and RNA structures into a 5 A-resolution map of the 50S ribosomal subunit.
Nature, 400, 1999
|
|
1C05
 
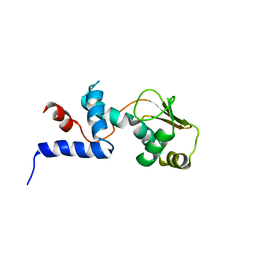 | | SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S4 DELTA 41, REFINED WITH DIPOLAR COUPLINGS (MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | RIBOSOMAL PROTEIN S4 DELTA 41 | | Authors: | Markus, M.A, Gerstner, R.B, Draper, D.E, Torchia, D.A. | | Deposit date: | 1999-07-14 | | Release date: | 1999-09-29 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Refining the overall structure and subdomain orientation of ribosomal protein S4 delta41 with dipolar couplings measured by NMR in uniaxial liquid crystalline phases.
J.Mol.Biol., 292, 1999
|
|
1C06
 
 | | SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S4 DELTA 41, REFINED WITH DIPOLAR COUPLINGS (ENSEMBLE OF 16 STRUCTURES) | | Descriptor: | RIBOSOMAL PROTEIN S4 DELTA 41 | | Authors: | Markus, M.A, Gerstner, R.B, Draper, D.E, Torchia, D.A. | | Deposit date: | 1999-07-14 | | Release date: | 1999-09-29 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Refining the overall structure and subdomain orientation of ribosomal protein S4 delta41 with dipolar couplings measured by NMR in uniaxial liquid crystalline phases.
J.Mol.Biol., 292, 1999
|
|
1C07
 
 | |
1C08
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV-HEN LYSOZYME COMPLEX | | Descriptor: | ANTI-HEN EGG WHITE LYSOZYME ANTIBODY (HYHEL-10), LYSOZYME | | Authors: | Shiroishi, M, Kondo, H, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 1999-07-15 | | Release date: | 2000-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of anti-Hen egg white lysozyme antibody (HyHEL-10) Fv-antigen complex. Local structural changes in the protein antigen and water-mediated interactions of Fv-antigen and light chain-heavy chain interfaces.
J.Biol.Chem., 274, 1999
|
|
1C09
 
 | | RUBREDOXIN V44A CP | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Min, T, Beard, B, Kang, C. | | Deposit date: | 1999-07-15 | | Release date: | 2001-02-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of the redox potential of the [Fe(SCys)(4)] site in rubredoxin by the orientation of a peptide dipole.
Biochemistry, 38, 1999
|
|
1C0A
 
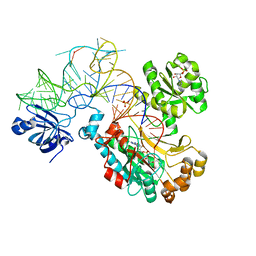 | | CRYSTAL STRUCTURE OF THE E. COLI ASPARTYL-TRNA SYNTHETASE : TRNAASP : ASPARTYL-ADENYLATE COMPLEX | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARTYL TRNA, ASPARTYL TRNA SYNTHETASE, ... | | Authors: | Eiler, S, Dock-Bregeon, A.-C, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-07-15 | | Release date: | 1999-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis of aspartyl-tRNA(Asp) in Escherichia coli--a snapshot of the second step.
EMBO J., 18, 1999
|
|
1C0B
 
 | |
1C0C
 
 | |
1C0E
 
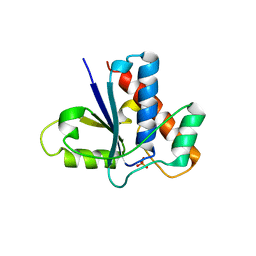 | | Active Site S19A Mutant of Bovine Heart Phosphotyrosyl Phosphatase | | Descriptor: | PHOSPHATE ION, PROTEIN (TYROSINE PHOSPHATASE (ORTHOPHOSPHORIC MONOESTER PHOSPHOHYDROLASE)) | | Authors: | Tabernero, L, Evans, B.N, Tishmack, P.A, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-07-15 | | Release date: | 1999-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the bovine protein tyrosine phosphatase dimer reveals a potential self-regulation mechanism.
Biochemistry, 38, 1999
|
|
1C0F
 
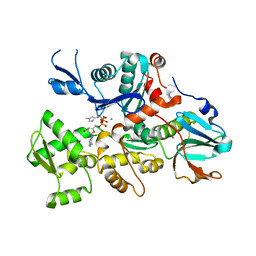 | | CRYSTAL STRUCTURE OF DICTYOSTELIUM CAATP-ACTIN IN COMPLEX WITH GELSOLIN SEGMENT 1 | | Descriptor: | ACTIN, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Matsuura, Y, Stewart, M, Kawamoto, M, Kamiya, N, Saeki, K, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 1999-07-16 | | Release date: | 2000-03-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the higher Ca(2+)-activation of the regulated actin-activated myosin ATPase observed with Dictyostelium/Tetrahymena actin chimeras
J.Mol.Biol., 296, 2000
|
|
1C0G
 
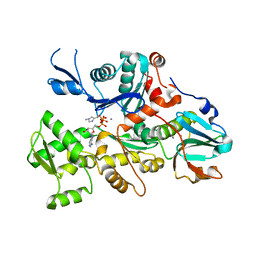 | | CRYSTAL STRUCTURE OF 1:1 COMPLEX BETWEEN GELSOLIN SEGMENT 1 AND A DICTYOSTELIUM/TETRAHYMENA CHIMERA ACTIN (MUTANT 228: Q228K/T229A/A230Y/E360H) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, PROTEIN (CHIMERIC ACTIN), ... | | Authors: | Matsuura, Y, Stewart, M, Kawamoto, M, Kamiya, N, Saeki, K, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 1999-07-16 | | Release date: | 2000-03-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the higher Ca(2+)-activation of the regulated actin-activated myosin ATPase observed with Dictyostelium/Tetrahymena actin chimeras.
J.Mol.Biol., 296, 2000
|
|
1C0I
 
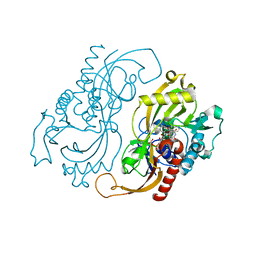 | | CRYSTAL STRUCTURE OF D-AMINO ACID OXIDASE IN COMPLEX WITH TWO ANTHRANYLATE MOLECULES | | Descriptor: | 2-AMINOBENZOIC ACID, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pollegioni, L, Diederichs, K, Molla, G, Umhau, S, Welte, W, Ghisla, S, Pilone, M.S. | | Deposit date: | 1999-07-16 | | Release date: | 2002-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Yeast d-amino Acid oxidase: structural basis of its catalytic properties
J.Mol.Biol., 324, 2002
|
|
1C0K
 
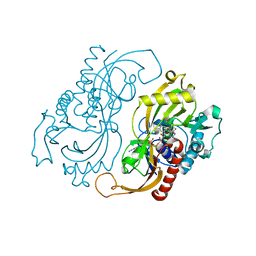 | | CRYSTAL STRUCTURE ANALYSIS OF D-AMINO ACID OXIDASE IN COMPLEX WITH L-LACTATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LACTIC ACID, PROTEIN (D-AMINO ACID OXIDASE) | | Authors: | Umhau, S, Molla, G, Diederichs, K, Pilone, M.S, Ghisla, S, Welte, W, Pollegioni, L. | | Deposit date: | 1999-07-16 | | Release date: | 2000-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C0L
 
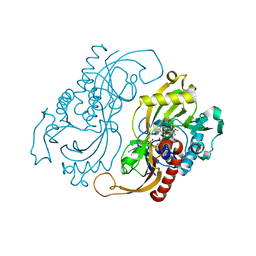 | | D-AMINO ACID OXIDASE: STRUCTURE OF SUBSTRATE COMPLEXES AT VERY HIGH RESOLUTION REVEAL THE CHEMICAL REACTTION MECHANISM OF FLAVIN DEHYDROGENATION | | Descriptor: | D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, TRIFLUOROALANINE | | Authors: | Umhau, S, Molla, G, Diederichs, K, Pilone, M.S, Ghisla, S, Welte, W. | | Deposit date: | 1999-07-16 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
