7CQU
 
 | | GmaS/ADP/MetSox-P complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQX
 
 | | GmaS/ADP complex-Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQW
 
 | | GmaS/ADP complex-Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
8HF3
 
 | | Cryo-EM structure of human ZDHHC9/GCP16 complex | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, Golgin subfamily A member 7, PALMITIC ACID, ... | | Authors: | Wu, J, Hu, Q, Zhang, Y, Liu, S, Yang, A. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Regulation of RAS palmitoyltransferases by accessory proteins and palmitoylation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HFC
 
 | | Cryo-EM structure of yeast Erf2/Erf4 complex | | Descriptor: | PALMITIC ACID, Palmitoyltransferase ERF2, Ras modification protein ERF4, ... | | Authors: | Wu, J, Hu, Q, Zhang, Y, Yang, A, Liu, S. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Regulation of RAS palmitoyltransferases by accessory proteins and palmitoylation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7XIC
 
 | | S-ECD (Omicron) in complex with STS165 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional analysis of an inter-Spike bivalent neutralizing antibody against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7XID
 
 | | S-ECD (Omicron) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional analysis of an inter-Spike bivalent neutralizing antibody against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7D6F
 
 | | The crystal structure of ARMS-PBM/MAGI2-PDZ4 | | Descriptor: | GLYCEROL, Kinase D-interacting substrate of 220 kDa, Membrane-associated guanylate kinase, ... | | Authors: | Ye, J, Zhang, Y, Zhong, Z, Wang, C. | | Deposit date: | 2020-09-30 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal structure of the PDZ4 domain of MAGI2 in complex with PBM of ARMS reveals a canonical PDZ recognition mode.
Neurochem.Int., 149, 2021
|
|
8I91
 
 | | ACE2-SIT1 complex bound with proline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
8I93
 
 | | ACE2-B0AT1 complex bound with methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
8I92
 
 | | ACE2-B0AT1 complex bound with glutamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
7XL4
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL3
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7VF9
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
8JE4
 
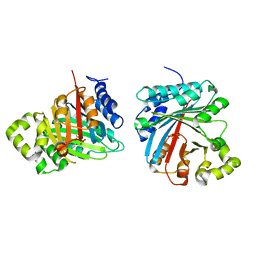 | | Crystal structure of LimF prenyltransferase (H239G/W273T mutant) bound with the thiodiphosphate moiety of farnesyl S-thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, TRIHYDROGEN THIODIPHOSPHATE, prenyltransferase, ... | | Authors: | Hamada, K, Oguni, A, Zhang, Y, Satake, M, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Switching Prenyl Donor Specificities of Cyanobactin Prenyltransferases.
J.Am.Chem.Soc., 145, 2023
|
|
7EH2
 
 | |
7EH1
 
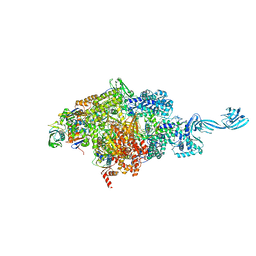 | | Thermus thermophilus transcription initiation complex containing a template-strand purine at position TSS-2, GpG RNA primer, and CMPcPP | | Descriptor: | 1,4-BUTANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (27-MER), ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Promoter-sequence determinants and structural basis of primer-dependent transcription initiation in Escherichia coli .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EGQ
 
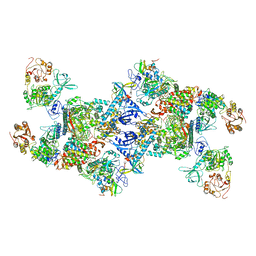 | | Co-transcriptional capping machineries in SARS-CoV-2 RTC: Coupling of N7-methyltransferase and 3'-5' exoribonuclease with polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L.M, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-03-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading.
Cell, 184, 2021
|
|
8W6D
 
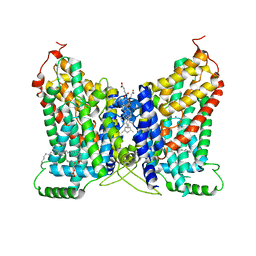 | | CryoEM structure of NaDC1 in apo state | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6O
 
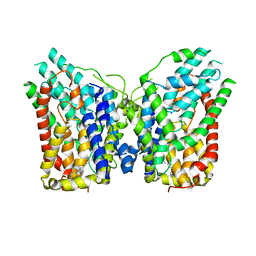 | | NaS1 in IN/IN state | | Descriptor: | CHOLESTEROL, SODIUM ION, Solute carrier family 13 member 1 | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6G
 
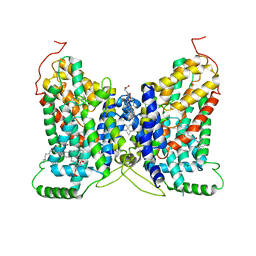 | | NaDC1 with inhibitor ACA | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-[[(~{E})-3-(4-pentylphenyl)prop-2-enoyl]amino]benzoic acid, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6C
 
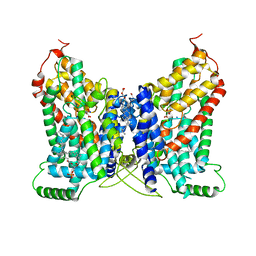 | | CryoEM structure of NaDC1 with Citrate | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CHOLESTEROL HEMISUCCINATE, CITRIC ACID, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Dai, L, Zhang, Y, Shen, Y, Chen, Y, Shi, T, Yang, H, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6T
 
 | | NaS1 in IN/OUT state | | Descriptor: | Solute carrier family 13 member 1 | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6H
 
 | | NaS1 with sulfate - IN/IN state | | Descriptor: | CHOLESTEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
7WQ4
 
 | | Galanin-bound galanin receptor 2 in complex with Gq | | Descriptor: | CHOLESTEROL, Engineered Guanine nucleotide-binding protein G(q) subunit alpha, Galanin, ... | | Authors: | Duan, J, Shen, D.D, Xu, H.E, Zhang, Y, Jiang, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-20 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis for allosteric agonism and G protein subtype selectivity of galanin receptors
Nat Commun, 13, 2022
|
|
