7WF9
 
 | |
7WF8
 
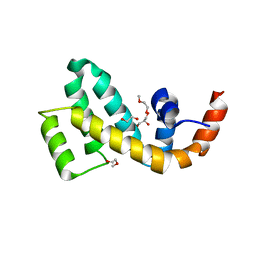 | | Crystal structure of mouse SNX25 RGS domain in space group P212121 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2021-12-26 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies Reveal Unique Non-canonical Regulators of G Protein Signaling Homology (RH) Domains in Sorting Nexins.
J.Mol.Biol., 434, 2022
|
|
7EU7
 
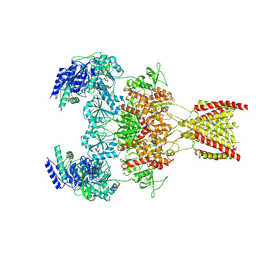 | | Structure of the human GluN1-GluN2A NMDA receptor in complex with S-ketamine, glycine and glutamate | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, Y, Zhang, T, Zhu, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-08-04 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of ketamine action on human NMDA receptors.
Nature, 596, 2021
|
|
8IHT
 
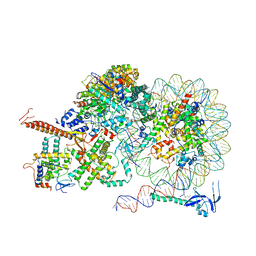 | | Rpd3S bound to the nucleosome | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, DNA (164-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHM
 
 | | Eaf3 CHD domain bound to the nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (164-MER), DNA (165-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHN
 
 | | Cryo-EM structure of the Rpd3S core complex | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, Histone H3, ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
7E1T
 
 | | Crystal structure of Rab9A-GTP-Nde1 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Isoform 2 of Nuclear distribution protein nudE homolog 1, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Zhang, T, Ding, J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Nde1 is a Rab9 effector for loading late endosomes to cytoplasmic dynein motor complex.
Structure, 30, 2022
|
|
7D0A
 
 | | Acinetobacter MlaFEDB complex in ADP-vanadate trapped Vclose conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-DIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D09
 
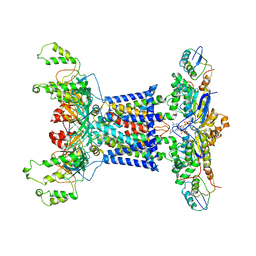 | | Acinetobacter MlaFEDB complex in ATP-bound Vtrans2 conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D08
 
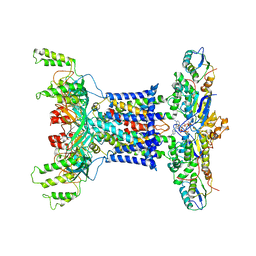 | | Acinetobacter MlaFEDB complex in ATP-bound Vtrans1 conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D06
 
 | | Cryo EM structure of the nucleotide free Acinetobacter MlaFEDB complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ABC transporter ATP-binding protein, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7F3L
 
 | |
7C8G
 
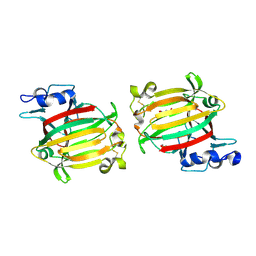 | | Structure of alginate lyase AlyC3 | | Descriptor: | Alginate lyase AlyC3, GLYCEROL, SUCCINIC ACID | | Authors: | Zhang, Y.Z, Xu, F, Chen, X.L, Wang, P. | | Deposit date: | 2020-05-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic.
J.Biol.Chem., 295, 2020
|
|
7C8F
 
 | | Structure of alginate lyase AlyC3 in complex with dimannuronate(2M) | | Descriptor: | H127A/Y244A mutant of alginate lyase AlyC3 in complex with dimannuronate, MALONATE ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Zhang, Y.Z, Xu, F, Chen, X.L, Wang, P. | | Deposit date: | 2020-05-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic.
J.Biol.Chem., 295, 2020
|
|
7F3I
 
 | |
7F3K
 
 | |
7F3J
 
 | |
7DK8
 
 | | Crystal structure of OsGH3-8 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Probable indole-3-acetic acid-amido synthetase GH3.8 | | Authors: | Zhang, Y.K, Xu, G.L, Ming, Z.H. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the acyl acid amido synthetase GH3-8 from Oryza sativa.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7E7S
 
 | | WT transporter state1 | | Descriptor: | CALCIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-02-27 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM analysis provides new mechanistic insight into ATP binding to Ca 2+ -ATPase SERCA2b.
Embo J., 40, 2021
|
|
7D6E
 
 | |
7D6D
 
 | |
4URT
 
 | | The crystal structure of a fragment of netrin-1 in complex with FN5- FN6 of DCC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Finci, L.I, Krueger, N, Sun, X, Zhang, J, Chegkazi, M, Wu, Y, Schenk, G, Mertens, H.D.T, Svergun, D.I, Zhang, Y, Wang, J.-h, Meijers, R. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Crystal Structure of Netrin-1 in Complex with Dcc Reveals the Bi-Functionality of Netrin-1 as a Guidance Cue
Neuron, 83, 2014
|
|
1VYU
 
 | | Beta3 subunit of Voltage-gated Ca2+-channel | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
5WTA
 
 | | Crystal Structure of Staphylococcus aureus SdrE apo form | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
3L0B
 
 | | Crystal structure of SCP1 phosphatase D206A mutant phosphoryl-intermediate | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1.
Protein Sci., 19, 2010
|
|
