7K75
 
 | |
7K76
 
 | |
3QG6
 
 | | Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody | | Descriptor: | AP4-24H11 Heavy Chain, AP4-24H11 Light Chain, Agr autoinducing peptide, ... | | Authors: | Kirchdoerfer, R.N, Janda, J.D, Kaufmann, G.F, Wilson, I.A. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Ligand Recognition and Discrimination of a Quorum-quenching Antibody.
J.Biol.Chem., 286, 2011
|
|
1EBA
 
 | |
5KUX
 
 | |
5KUY
 
 | |
5FEH
 
 | | Crystal structure of PCT64_35B, a broadly neutralizing anti-HIV antibody | | Descriptor: | 1,2-ETHANEDIOL, PCT64_26 Fab heavy chain, PCT64_26 Fab light chain, ... | | Authors: | Murrell, S, Wilson, I.A. | | Deposit date: | 2015-12-17 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | HIV Envelope Glycoform Heterogeneity and Localized Diversity Govern the Initiation and Maturation of a V2 Apex Broadly Neutralizing Antibody Lineage.
Immunity, 47, 2017
|
|
1ERN
 
 | |
3Q1S
 
 | | HIV-1 neutralizing antibody Z13e1 in complex with epitope display protein | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Interleukin-22, ... | | Authors: | Stanfield, R.L, Julien, J.-P, Pejchal, R, Gach, J.S, Zwick, M.B, Wilson, I.A. | | Deposit date: | 2010-12-17 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of a Protein Immunogen that Displays an HIV-1 gp41 Neutralizing Epitope.
J.Mol.Biol., 414, 2011
|
|
3OZ9
 
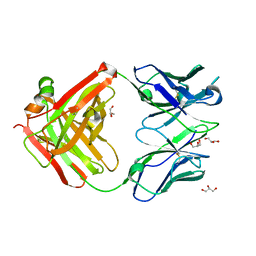 | |
7JMO
 
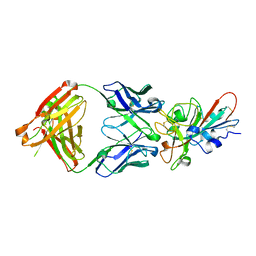 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-04 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-04 heavy chain, COVA2-04 light chain, ... | | Authors: | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
3QQB
 
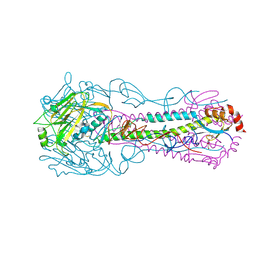 | | Crystal structure of HA2 R106H mutant of H2 hemagglutinin, neutral pH form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Characterization of an Early Fusion Intermediate of Influenza Virus Hemagglutinin.
J.Virol., 85, 2011
|
|
5ILT
 
 | | Crystal structure of bovine Fab A01 | | Descriptor: | bovine Fab A01 heavy chain, bovine Fab A01 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-03-04 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
7KN5
 
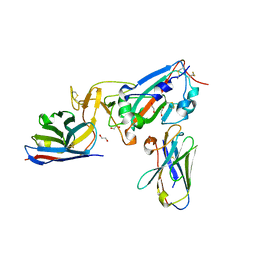 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobodies VHH E and U | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
1F58
 
 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 24-RESIDUE PEPTIDE (RESIDUES 308-333 OF HIV-1 GP120 (MN ISOLATE) WITH ALA TO AIB SUBSTITUTION AT POSITION 323 | | Descriptor: | Envelope glycoprotein gp120, PROTEIN (IGG1 ANTIBODY 58.2 (HEAVY CHAIN)), PROTEIN (IGG1 ANTIBODY 58.2 (LIGHT CHAIN)) | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-21 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
5JOR
 
 | | Crystal structure of unbound anti-glycan antibody Fab14.22 at 2.2 A | | Descriptor: | Fab 14.22 light chain, Fab14.22 heavy chain, GLYCEROL, ... | | Authors: | Sarkar, A, Irimia, A, Teyton, L, Wilson, I.A. | | Deposit date: | 2016-05-02 | | Release date: | 2017-03-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | T cells control the generation of nanomolar-affinity anti-glycan antibodies.
J. Clin. Invest., 127, 2017
|
|
5IJV
 
 | | Crystal structure of bovine Fab E03 | | Descriptor: | bovine Fab E03 heavy chain, bovine Fab E03 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-03-02 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
3MJ6
 
 | |
5IF0
 
 | |
3RG1
 
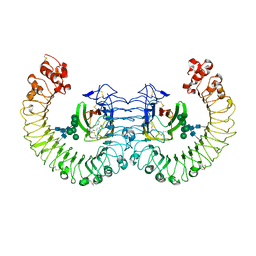 | | Crystal structure of the RP105/MD-1 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CD180 molecule, LY86 protein, ... | | Authors: | Yoon, S.I, Hong, M, Wilson, I.A. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | An unusual dimeric structure and assembly for TLR4 regulator RP105-MD-1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
5HZQ
 
 | | Crystal structure of cellular retinoic acid binding protein 2 (CRABP2)-aryl fluorosulfate covalent conjugate | | Descriptor: | 4'-[(3,6,9,12-tetraoxapentadec-14-yn-1-yl)oxy][1,1'-biphenyl]-4-yl sulfurofluoridate, Cellular retinoic acid-binding protein 2, GLYCEROL | | Authors: | Chen, W, Mortenson, D.E, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2016-02-02 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue.
J.Am.Chem.Soc., 138, 2016
|
|
3R06
 
 | |
7KN7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobody VHH W and antibody Fab CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, CC12.3 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7KN6
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobody VHH V and antibody Fab CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, CC12.3 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
3QQO
 
 | |
