6SU9
 
 | |
6SKS
 
 | |
6SKT
 
 | |
5CB2
 
 | | the structure of candida albicans Sey1p in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein SEY1 | | Authors: | Yan, L, Sun, S, Wang, W, Shi, J, Hu, X, Wang, S, Rao, Z, Hu, J, Lou, Z. | | Deposit date: | 2015-06-30 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion
J.Cell Biol., 210, 2015
|
|
3NBI
 
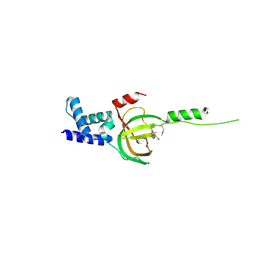 | | Crystal structure of human RMI1 N-terminus | | Descriptor: | RecQ-mediated genome instability protein 1 | | Authors: | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
3NBH
 
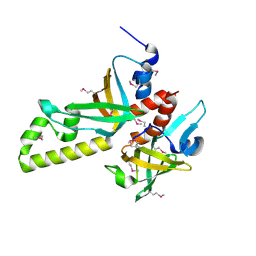 | | Crystal structure of human RMI1C-RMI2 complex | | Descriptor: | RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | Authors: | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
4MGR
 
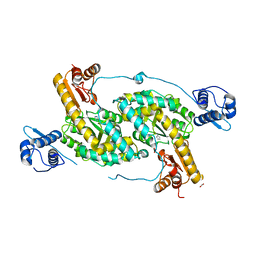 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5BMS
 
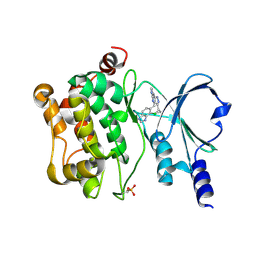 | |
4WQ6
 
 | | The crystal structure of human Nicotinamide phosphoribosyltransferase (NAMPT) in complex with N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide inhibitor (compound 21) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Li, D, Wang, W. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Identification of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with no evidence of CYP3A4 time-dependent inhibition and improved aqueous solubility.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5BKF
 
 | | Cyro-EM structure of human Glycine Receptor alpha2-beta heteromer, Glycine bound, desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alpha-2, ... | | Authors: | Yu, H, Wang, W. | | Deposit date: | 2021-03-19 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Characterization of the subunit composition and structure of adult human glycine receptors
Neuron, 109, 2021
|
|
5BKG
 
 | | Cyro-EM structure of human Glycine Receptor alpha2-beta heteromer, glycine bound, (semi)open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alpha-2, ... | | Authors: | Yu, H, Wang, W. | | Deposit date: | 2021-03-19 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Characterization of the subunit composition and structure of adult human glycine receptors
Neuron, 109, 2021
|
|
5KQA
 
 | | Crystal structure of buckwheat glutaredoxin-glutathione complex | | Descriptor: | GLUTATHIONE, Glutaredoxin-glutathione complex | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
9BIK
 
 | | Crystal structure of inhibitor 1 bound to HPK1 | | Descriptor: | (1S,2S)-N-[(6P)-8-amino-6-(4-methylpyridin-3-yl)isoquinolin-3-yl]-2-cyanocyclopropane-1-carboxamide, 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Kiefer, J.T, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Siu, M, Heffron, T.P, Choo, E.F. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
9B3B
 
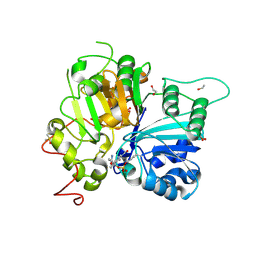 | | Structure of TDP1 complexed with compound IB09 | | Descriptor: | (8M)-8-{4-(benzylcarbamoyl)-2-[(fluorosulfonyl)oxy]phenyl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke, T.R. | | Deposit date: | 2024-03-18 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of TDP1 complexed with compound IB09
To Be Published
|
|
9BJ1
 
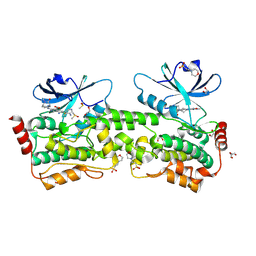 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (4S,5R,7R,11aP)-10-{[(3R)-3-hydroxy-1-methyl-2-oxopyrrolidin-3-yl]ethynyl}-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, (9S)-2-{[(6P)-8-amino-6-(5-amino-4-methylpyridin-3-yl)-7-fluoroisoquinolin-3-yl]amino}-6-methyl-5,6-dihydro-4H-pyrazolo[1,5-d][1,4]diazepin-7(8H)-one, 1,2-ETHANEDIOL, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-24 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
9BI8
 
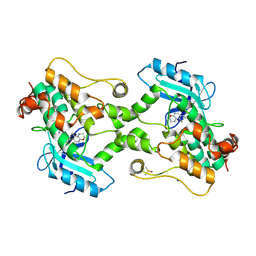 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (3R,4S)-4-methyloxolan-3-yl [(6P)-8-amino-7-fluoro-6-(8-methyl-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)isoquinolin-3-yl]carbamate, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase kinase kinase kinase 1, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-22 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
1G6M
 
 | |
1JE9
 
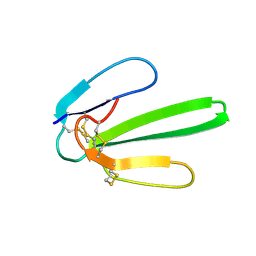 | | NMR SOLUTION STRUCTURE OF NT2 | | Descriptor: | SHORT NEUROTOXIN II | | Authors: | Cheng, Y, Wang, W, Wang, J. | | Deposit date: | 2001-06-16 | | Release date: | 2001-07-04 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of three neurotoxins from the venom of Naja kaouthia: a comparison between the NMR-derived structure of NT2 with its homologues, NT1 and NT3
BIOCHIM.BIOPHYS.ACTA, 1594, 2002
|
|
4XZC
 
 | | The crystal structure of Kupe virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZE
 
 | | The crystal structure of Hazara virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZA
 
 | | The crystal structure of Erve virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZ8
 
 | | The crystal structure of Erve virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
5KIT
 
 | |
1DCH
 
 | | CRYSTAL STRUCTURE OF DCOH, A BIFUNCTIONAL, PROTEIN-BINDING TRANSCRIPTION COACTIVATOR | | Descriptor: | DCOH (DIMERIZATION COFACTOR OF HNF-1), SULFATE ION | | Authors: | Endrizzi, J.A, Cronk, J.D, Wang, W, Crabtree, G.R, Alber, T. | | Deposit date: | 1995-01-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of DCoH, a bifunctional, protein-binding transcriptional coactivator.
Science, 268, 1995
|
|
1FRY
 
 | | THE SOLUTION STRUCTURE OF SHEEP MYELOID ANTIMICROBIAL PEPTIDE, RESIDUES 1-29 (SMAP29) | | Descriptor: | MYELOID ANTIMICROBIAL PEPTIDE | | Authors: | Tack, B.F, Sawai, M.V, Kearney, W.R, Robertson, A.D, Sherman, M.A, Wang, W, Hong, T, Boo, L.M, Wu, H, Waring, A.J, Lehrer, R.I. | | Deposit date: | 2000-09-07 | | Release date: | 2002-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | SMAP-29 has two LPS-binding sites and a central hinge.
Eur.J.Biochem., 269, 2002
|
|
