7EVM
 
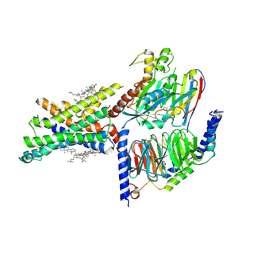 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
2N62
 
 | | ddFLN5+110 | | Descriptor: | gelation factor, secretion monitor chimera | | Authors: | Cabrita, L.D, Cassaignau, A.M.E, Launay, H.M.M, Waudby, C.A, Camilloni, C, Robertson, A.L, Wang, X, Wlodarski, T, Wentink, A.S, Vendruscolo, M, Dobson, C.M, Christodoulou, J. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2PQ6
 
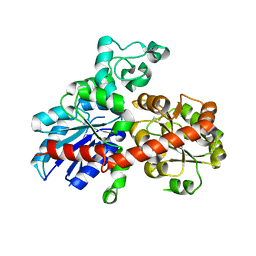 | |
8GXQ
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in MH-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X, Xu, Y. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (5.04 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
8GXS
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in H-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
5ZMY
 
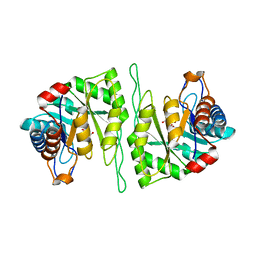 | | Crystal structure of a cis-epoxysuccinate hydrolase producing D(-)-tartaric acids | | Descriptor: | Cis-epoxysuccinate hydrolase, D(-)-TARTARIC ACID, ZINC ION | | Authors: | Dong, S, Liu, X, Wang, X, Feng, Y. | | Deposit date: | 2018-04-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure d(-)-tartaric acid.
Chem. Commun. (Camb.), 54, 2018
|
|
7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7D7N
 
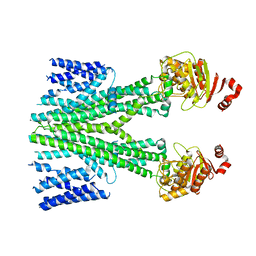 | | Cryo-EM structure of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
7D7R
 
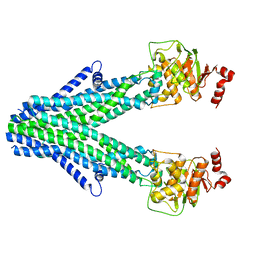 | | Cryo-EM structure of the core domain of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
7F4G
 
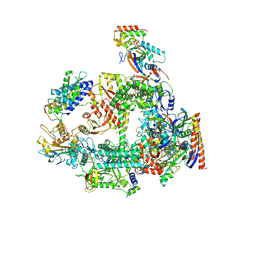 | | Structure of RPAP2-bound RNA polymerase II | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2021-07-07 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | RPAP2 regulates a transcription initiation checkpoint by inhibiting assembly of pre-initiation complex.
Cell Rep, 39, 2022
|
|
5ZMU
 
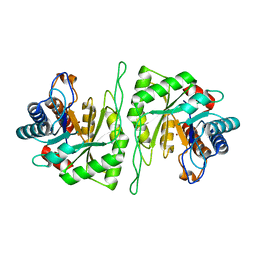 | | Crystal structure of a cis-epoxysuccinate hydrolase producing D(-)-tartaric acids | | Descriptor: | Cis-epoxysuccinate hydrolase, ZINC ION | | Authors: | Dong, S, Liu, X, Wang, X, Feng, Y. | | Deposit date: | 2018-04-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure d(-)-tartaric acid.
Chem. Commun. (Camb.), 54, 2018
|
|
7FAE
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2(state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P36-5D2 heavy chain, ... | | Authors: | Zhang, L, Wang, X, Shan, S, Zhang, S. | | Deposit date: | 2021-07-06 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7FAF
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2 (state1) | | Descriptor: | P36-5D2 heavy chain, P36-5D2 light chain, Spike glycoprotein | | Authors: | Zhang, L, Wang, X, Zhang, S, Shan, S. | | Deposit date: | 2021-07-06 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7C9W
 
 | | E30 F-particle in complex with CD55 | | Descriptor: | Complement decay-accelerating factor, MYRISTIC ACID, SPHINGOSINE, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9U
 
 | | Echovirus 30 E-particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9V
 
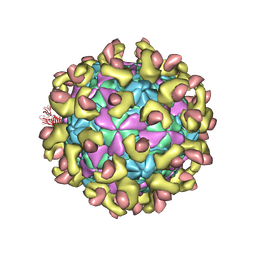 | | E30 F-particle in complex with FcRn | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, MYRISTIC ACID, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C81
 
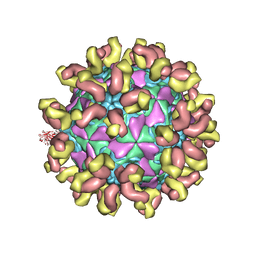 | | E30 F-particle in complex with 6C5 | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Wang, K, Zheng, B, Zhang, L, Cui, L, Su, X, Zhang, Q, Guo, Y, Zhu, L, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
7C9S
 
 | | Echovirus 30 F-particle | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Wang, K, Sun, Y, Zhu, L, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9T
 
 | | Echovirus 30 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C80
 
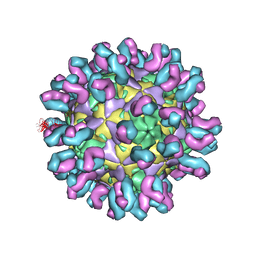 | | E30 F-particle in complex with 4B10 | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Wang, K, Zheng, B, Zhang, L, Cui, L, Su, X, Zhang, Q, Guo, Y, Zhu, L, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
7C9Z
 
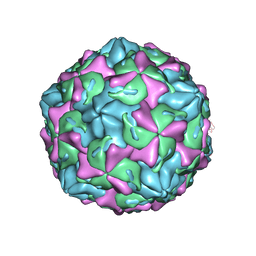 | | Coxsackievirus B1 F-particle | | Descriptor: | MYRISTIC ACID, PALMITIC ACID, VP1, ... | | Authors: | Feng, R, Wang, K, Rao, Z, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
6I2K
 
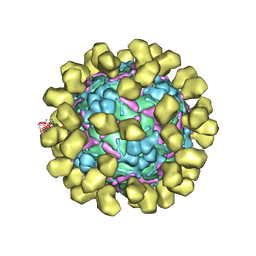 | | Structure of EV71 complexed with its receptor SCARB2 | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Zhao, Y, Kotecha, A, Fry, E.E, Kelly, J, Wang, X, Rao, Z, Rowlands, D.J, Ren, J, Stuart, D.I. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2.
Nat Microbiol, 4, 2019
|
|
6JHR
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F6 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHT
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F9 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
