7WTK
 
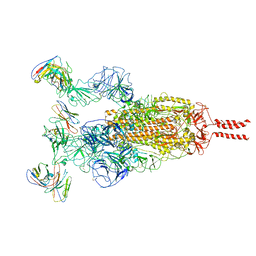 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv286 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv286, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTF
 
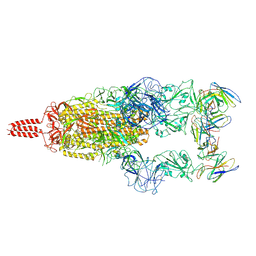 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv051 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv051, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTG
 
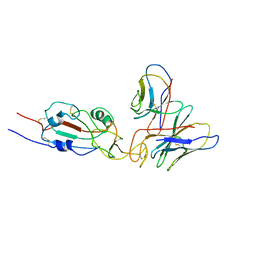 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv051 | | Descriptor: | Heavy chain of XGv051, Light chain of XGv051, Spike protein S1 | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTH
 
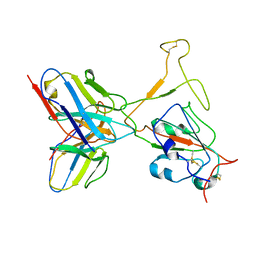 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv264 | | Descriptor: | Heavy chain of XGv264, Light chain of XGv264, Spike protein S1 | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTI
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv264 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv264, Light chain of XGv264, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
8YM2
 
 | | Crystal structure of AIDA-1 PTB domain in complex with SynGAP NPxF motif | | Descriptor: | Ankyrin repeat and sterile alpha motif domain-containing protein 1B, Ras/Rap GTPase-activating protein SynGAP | | Authors: | Wang, X, Wang, Y, Cai, Q, Zhang, M. | | Deposit date: | 2024-03-08 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AIDA-1/ANKS1B Binds to the SynGAP Family RasGAPs with High Affinity and Specificity.
J.Mol.Biol., 436, 2024
|
|
7CWU
 
 | | SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
8Y3Q
 
 | | ASFV p72 in complex with Fab F11 | | Descriptor: | B646L, Heavy chain of F11, Light chain of F11 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8Y3O
 
 | | ASFV p72 in complex with Fab B1 | | Descriptor: | B646L, Heavy chain of B1, Light chain of B1 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8Y3R
 
 | | ASFV p72 in complex with Fab H3 | | Descriptor: | B646L, Heavy chain of H3, Light chain of H3 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8Y3P
 
 | | ASFV p72 in complex with Fab C9 | | Descriptor: | B646L, Heavy chain of C9, Light chain of C9 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
2GMG
 
 | | Solution NMR Structure of protein PF0610 from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfG3 | | Descriptor: | hypothetical protein Pf0610 | | Authors: | Wang, X, Lee, H.S, Adams, M.W, Northeast Structural Genomics Consortium (NESG), Montelione, G.T, Prestegard, J.H. | | Deposit date: | 2006-04-06 | | Release date: | 2006-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PF0610, a novel winged helix-turn-helix variant possessing a rubredoxin-like Zn ribbon motif from the hyperthermophilic archaeon, Pyrococcus furiosus.
Biochemistry, 46, 2007
|
|
8GX9
 
 | |
7WT9
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab 9A8 | | Descriptor: | Heavy chain of Fab 9A8, Light chain of Fab 9A8, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7WT7
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab 9A8 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 9A8, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7WT8
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab 9A8 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 9A8, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
2KJK
 
 | | Solution structure of the second domain of the listeria protein Lin2157, Northeast Structural Genomics Consortium target Lkr136b | | Descriptor: | Lin2157 protein | | Authors: | Wang, X, Hamilton, K, Xiao, R.H, Lee, D, Ciccosanti, C.H, Nair, R, Rost, B, Acton, T.B, Swapna, G, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-29 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Lkr136b
To be Published
|
|
2K7N
 
 | |
2JT3
 
 | | Solution Structure of F153W cardiac troponin C | | Descriptor: | Troponin C | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-18 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
2JTZ
 
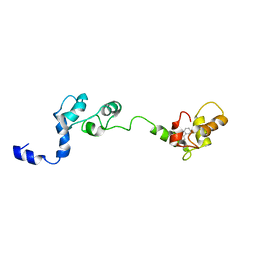 | | Solution structure and chemical shift assignments of the F104-to-5-flurotryptophan mutant of cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-28 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
2JZC
 
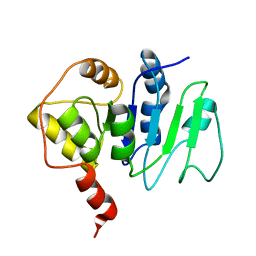 | | NMR solution structure of ALG13: The sugar donor subunit of a yeast N-acetylglucosamine transferase. Northeast Structural Genomics Consortium target YG1 | | Descriptor: | UDP-N-acetylglucosamine transferase subunit ALG13 | | Authors: | Wang, X, Weldeghorghis, T, Zhang, G, Imepriali, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Alg13: the sugar donor subunit of a yeast N-acetylglucosamine transferase.
Structure, 16, 2008
|
|
8ZL9
 
 | | ASFV p72 in complex with Fab G6 | | Descriptor: | B646L, G6 Heavy chain, G6 Light chain | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-05-17 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
2JT8
 
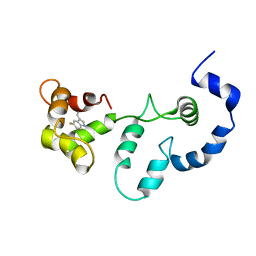 | | Solution structure of the F153-to-5-flurotryptophan mutant of human cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
2K2R
 
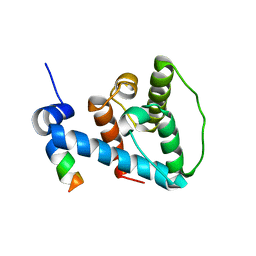 | | The NMR structure of alpha-parvin CH2/paxillin LD1 complex | | Descriptor: | Alpha-parvin, Paxillin | | Authors: | Wang, X, Fukuda, K, Byeon, I, Velyvis, A, Wu, C, Gronenborn, A, Qin, J. | | Deposit date: | 2008-04-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of {alpha}-Parvin CH2-Paxillin LD1 Complex Reveals a Novel Modular Recognition for Focal Adhesion Assembly.
J.Biol.Chem., 283, 2008
|
|
7FEE
 
 | | Crystal structure of the allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, ... | | Authors: | Wang, X, Zhao, C, Shao, Z. | | Deposit date: | 2021-07-19 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
