8TPK
 
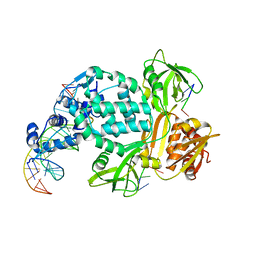 | | P6522 crystal form of C. crescentus DriD-ssDNA-DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*TP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*C)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structure of the WYL-domain containing transcription activator, DriD, in complex with ssDNA effector and DNA target site.
Nucleic Acids Res., 52, 2024
|
|
2GJE
 
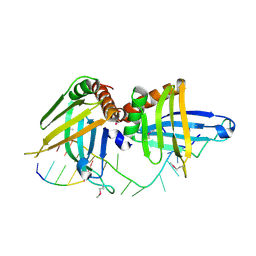 | | Structure of a guideRNA-binding protein complex bound to a gRNA | | Descriptor: | RNA tetramer, guide RNA 40-mer, mitochondrial RNA-binding protein 1, ... | | Authors: | Schumacher, M.A, Karamooz, E, Zikova, A, Trantirek, L, Lukes, J. | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism.
Cell(Cambridge,Mass.), 126, 2006
|
|
8UFJ
 
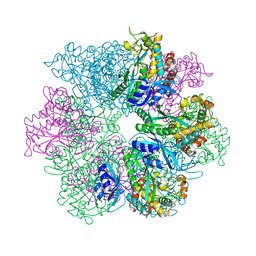 | | Structure of M. mazei GS(R167L-A168G) apo form | | Descriptor: | Glutamine synthetase, MAGNESIUM ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-10-04 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | M. mazei glutamine synthetase and glutamine synthetase-GlnK1 structures reveal enzyme regulation by oligomer modulation.
Nat Commun, 14, 2023
|
|
2G66
 
 | |
5U1J
 
 | | Structure of pNOB8 ParA bound to nonspecific DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*TP*AP*AP*TP*GP*AP*CP*GP*CP*CP*GP*GP*CP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*GP*CP*CP*GP*GP*CP*GP*TP*CP*AP*TP*GP*AP*CP*AP*CP*G)-3'), PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-04-19 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of partition protein ParA with nonspecific DNA and ParB effector reveal molecular insights into principles governing Walker-box DNA segregation.
Genes Dev., 31, 2017
|
|
6UEP
 
 | |
5U1G
 
 | | Structure of TP228 ParA-AMPPNP-ParB complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ParA, TP228 ParB fragment | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structures of partition protein ParA with nonspecific DNA and ParB effector reveal molecular insights into principles governing Walker-box DNA segregation.
Genes Dev., 31, 2017
|
|
5TZF
 
 | | Structure of the BldD CTD(D116A)-(c-di-GMP)2 intermediate, form 1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
5TZG
 
 | | Structure of the BldD CTD(D116A)-(c-di-GMP)2, form 2 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein, ZINC ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
6UEO
 
 | | Structure of A. thaliana TBP-AC mismatch DNA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*CP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
1DBR
 
 | | HYPOXANTHINE GUANINE XANTHINE | | Descriptor: | HYPOXANTHINE GUANINE XANTHINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION | | Authors: | Schumacher, M.A, Carter, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1996-02-13 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Toxoplasma gondii HGXPRTase reveal the catalytic role of a long flexible loop.
Nat.Struct.Biol., 3, 1996
|
|
6UER
 
 | |
1DH3
 
 | |
6UEQ
 
 | | Structure of TBP bound to C-C mismatch containing TATA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*CP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*C)-3'), SULFATE ION, ... | | Authors: | Schumacher, M.A, Al-Hashimi, H. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
1ZX4
 
 | | Structure of ParB bound to DNA | | Descriptor: | CITRIC ACID, Plasmid Partition par B protein, parS-small DNA centromere site | | Authors: | Schumacher, M.A, Funnell, B.E. | | Deposit date: | 2005-06-06 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structures of ParB bound to DNA reveal mechanism of partition complex formation.
Nature, 438, 2005
|
|
4YJ1
 
 | | Crystal structure of T. brucei MRB1590-ADP bound to poly-U RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2015-03-03 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the T. brucei kRNA editing factor MRB1590 reveal unique RNA-binding pore motif contained within an ABC-ATPase fold.
Nucleic Acids Res., 43, 2015
|
|
2Q2K
 
 | | Structure of nucleic-acid binding protein | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*AP*GP*TP*AP*TP*AP*(5IU)P*AP*CP*(5IU)P*AP*GP*TP*AP*TP*AP*TP*AP*CP*T)-3'), Hypothetical protein | | Authors: | Schumacher, M.A, Glover, T, Firth, N. | | Deposit date: | 2007-05-28 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Segrosome structure revealed by a complex of ParR with centromere DNA.
Nature, 450, 2007
|
|
1ZVV
 
 | | Crystal structure of a ccpa-crh-dna complex | | Descriptor: | DNA recognition strand CRE, Glucose-resistance amylase regulator, HPr-like protein crh, ... | | Authors: | Schumacher, M.A, Brennan, R.G, Hillen, W, Seidel, G. | | Deposit date: | 2005-06-02 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.
J.Biol.Chem., 281, 2006
|
|
1UPF
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V BOUND TO THE DRUG 5-FLUOROURACIL | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-06-17 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1UPU
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V, BOUND TO PRODUCT URIDINE-1-MONOPHOSPHATE (UMP) | | Descriptor: | PHOSPHATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-04-16 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1KQ1
 
 | | 1.55 A Crystal structure of the pleiotropic translational regulator, Hfq | | Descriptor: | ACETIC ACID, Host Factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
1KQ2
 
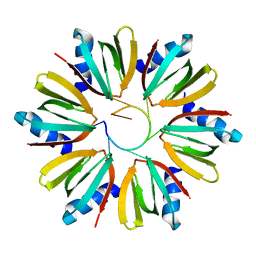 | | Crystal Structure of an Hfq-RNA Complex | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*G)-3', Host factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
6CG8
 
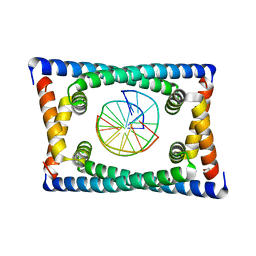 | | Structure of C. crescentus GapR-DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*AP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*TP*TP*TP*AP*A)-3'), UPF0335 protein B7Z12_12435 | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication.
Cell, 175, 2018
|
|
6CFX
 
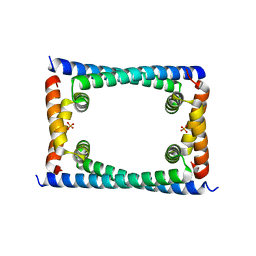 | | Bosea sp GapR solved in the presence of DNA | | Descriptor: | PHOSPHATE ION, UPF0335 protein ASE63_04290 | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-02-18 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication.
Cell, 175, 2018
|
|
1YM8
 
 | |
