4FC9
 
 | | Structure of the C-terminal domain of the type III effector Xcv3220 (XopL) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, uncharacterized protein | | Authors: | Singer, A.U, Xu, X, Cui, H, Tan, K, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminal domain of the type III effector Xcv3220 (XopL)
To be Published
|
|
3CNV
 
 | | Crystal structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica | | Descriptor: | CHLORIDE ION, CITRATE ANION, Putative GntR-family transcriptional regulator | | Authors: | Zimmerman, M.D, Xu, X, Cui, H, Filippova, E.V, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica.
To be Published
|
|
2PZ9
 
 | | Crystal structure of putative transcriptional regulator SCO4942 from Streptomyces coelicolor | | Descriptor: | Putative regulatory protein, SULFATE ION | | Authors: | Filippova, E.V, Chruszcz, M, Xu, X, Zheng, H, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-17 | | Release date: | 2007-06-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In situ proteolysis for protein crystallization and structure determination.
Nat.Methods, 4, 2007
|
|
1Y8A
 
 | | Structure of gene product AF1437 from Archaeoglobus fulgidus | | Descriptor: | MAGNESIUM ION, hypothetical protein AF1437 | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of gene product AF1437 from Archaeoglobus fulgidus
To be published
|
|
3CZQ
 
 | | Crystal structure of putative polyphosphate kinase 2 from Sinorhizobium meliloti | | Descriptor: | FORMIC ACID, GLYCEROL, Putative polyphosphate kinase 2 | | Authors: | Osipiuk, J, Evdokimova, E, Nocek, B, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3UDU
 
 | | Crystal structure of putative 3-isopropylmalate dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydrogenase, CHLORIDE ION | | Authors: | Tkaczuk, K.L, Chruszcz, M, Grimshaw, S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-28 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative 3-isopropylmalate dehydrogenase from Campylobacter jejuni
To be Published
|
|
6M8V
 
 | | Crystal structure of UbiX-like FMN prenyltransferase MJ0101 from Methanocaldococcus jannaschii, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Khusnutdinova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase MJ0101 from Methanocaldococcus jannaschii, FMN complex
To Be Published
|
|
3DCL
 
 | | Crystal structure of TM1086 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Chruszcz, M, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of TM1086
To be Published
|
|
3CKD
 
 | | Crystal structure of the C-terminal domain of the Shigella type III effector IpaH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Invasion plasmid antigen, ... | | Authors: | Lam, R, Singer, A.U, Cuff, M.E, Skarina, T, Kagan, O, DiLeo, R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2AZP
 
 | | Crystal Structure of PA1268 Solved by Sulfur SAD | | Descriptor: | hypothetical protein PA1268 | | Authors: | Liu, Y, Gorodichtchenskaia, E, Skarina, T, Yang, C, Joachimiak, A, Edwards, A, Pai, E.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of PA1268 Solved by Sulfur SAD
To be Published
|
|
4DFU
 
 | | Inhibition of an antibiotic resistance enzyme: crystal structure of aminoglycoside phosphotransferase APH(2")-ID/APH(2")-IVA in complex with kanamycin inhibited with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, APH(2")-Id, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Egorova, E, Di Leo, R, Li, H, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-24 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
3DCA
 
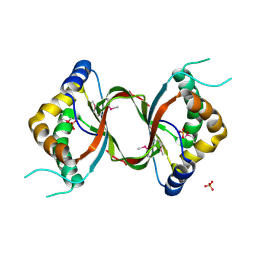 | | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target | | Descriptor: | RPA0582, SULFATE ION | | Authors: | Sledz, P, Wang, S, Chruszcz, M, Yim, V, Kudritska, M, Evdokimova, E, Turk, D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target
To be Published
|
|
1Y0U
 
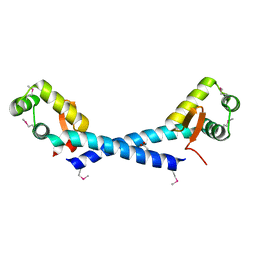 | | Crystal Structure of the putative arsenical resistance operon repressor from Archaeoglobus fulgidus | | Descriptor: | ACETATE ION, arsenical resistance operon repressor, putative | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-16 | | Release date: | 2004-12-28 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the putative arsenical resistance operon repressor from Archaeoglobus fulgidus
To be Published
|
|
4EHJ
 
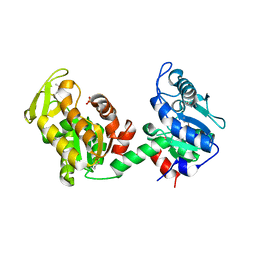 | | An X-ray Structure of a Putative Phosphogylcerate Kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Phosphoglycerate kinase, SULFATE ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Gordon, E, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | An X-ray Structure of a Putative Phosphogylcerate Kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4EVU
 
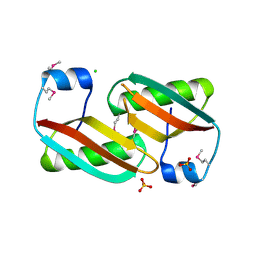 | | Crystal structure of C-terminal domain of putative periplasmic protein ydgH from S. enterica | | Descriptor: | CHLORIDE ION, Putative periplasmic protein ydgH, SULFATE ION | | Authors: | Michalska, K, Cui, H, Xu, X, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Savchenko, A, Adkins, J.N, Joachimiak, A, Program for the Characterization of Secreted Effector Proteins (PCSEP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-30 | | Last modified: | 2014-07-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
6BVC
 
 | | Crystal structure of AAC(3)-Ia in complex with coenzyme A | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside-(3)-N-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-12 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | To be published
To Be Published
|
|
3BQY
 
 | | Crystal structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2). | | Descriptor: | ACETIC ACID, PHOSPHATE ION, Putative TetR family transcriptional regulator | | Authors: | Cuff, M.E, Skarina, T, Kagan, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2).
TO BE PUBLISHED
|
|
4FCG
 
 | | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Zimmerman, M.D, Minor, W, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-13 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL)
To be Published
|
|
6M8T
 
 | | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX, PHOSPHATE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutidnova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex
To Be Published
|
|
3DR6
 
 | | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, yncA | | Authors: | Singer, A.U, Skarina, T, Onopriyenko, O, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-10 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Funded by the national institute of
allergy and infectious diseases of nih (contract number
hhsn272200700058c).
To be Published
|
|
6MIJ
 
 | | Crystal structure of EF-Tu from Acinetobacter baumannii in complex with Mg2+ and GDP | | Descriptor: | Elongation factor Tu, FORMIC ACID, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-19 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | To be published
To Be Published
|
|
6MN2
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA
To Be Published
|
|
6MR3
 
 | | Crystal structure of the competence-damaged protein (CinA) superfamily protein from Streptococcus mutans | | Descriptor: | CHLORIDE ION, Putative competence-damage inducible protein | | Authors: | Stogios, P.J, Cuff, M, Xu, X, Cui, H, Di Leo, R, Yim, V, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-10-11 | | Release date: | 2018-10-24 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the competence-damaged protein (CinA) superfamily protein from Streptococcus mutans
To Be Published
|
|
3CNI
 
 | | Crystal structure of a domain of a putative ABC type-2 transporter from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Putative ABC type-2 transporter | | Authors: | Filippova, E.V, Shumilin, I, Tkaczuk, K.L, Cymborowski, M, Chruszcz, M, Xu, X, Que, Q, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the putative ABC-type 2 transporter from Thermotoga maritima MSB8.
J.Struct.Funct.Genom., 15, 2014
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
