2NP3
 
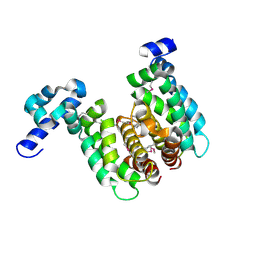 | | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3. | | Descriptor: | Putative TetR-family regulator | | Authors: | Koclega, K.D, Xu, X, Chruszcz, M, Gu, J, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3.
To be Published
|
|
3QSL
 
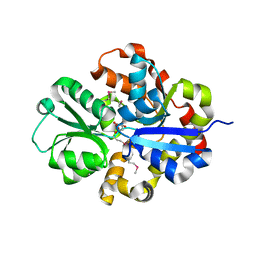 | | Structure of CAE31940 from Bordetella bronchiseptica RB50 | | Descriptor: | CITRIC ACID, Putative exported protein | | Authors: | Bajor, J, Kagan, O, Chruszcz, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of pyrimidine/thiamin biosynthesis precursor-like domain-containing protein CAE31940 from proteobacterium Bordetella bronchiseptica RB50, and evolutionary insight into the NMT1/THI5 family.
J Struct Funct Genomics, 15, 2014
|
|
2NNN
 
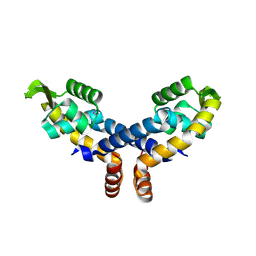 | | Crystal structure of probable transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator | | Authors: | Chang, C, Evdokimova, E, Altamentova, S, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-24 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of probable transcriptional regulator from Pseudomonas aeruginosa
To be Published
|
|
3Q7H
 
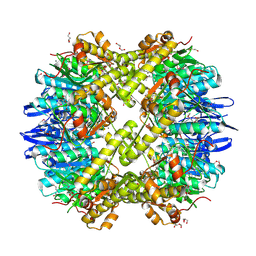 | | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, DI(HYDROXYETHYL)ETHER | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii
To be Published
|
|
3HFU
 
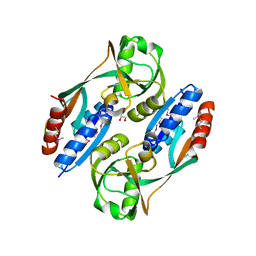 | | Crystal structure of the ligand binding domain of E. coli CynR with its specific effector azide | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, HTH-type transcriptional regulator cynR | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Dong, A, Edwards, A.M, Savchenko, A. | | Deposit date: | 2009-05-12 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the ligand binding domain of E. coli CynR with its specific effector azide
TO BE PUBLISHED
|
|
3HHQ
 
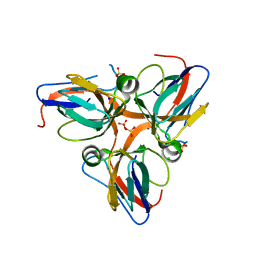 | | Crystal structure of apo dUT1p from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Dong, A, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2009-05-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
2QKO
 
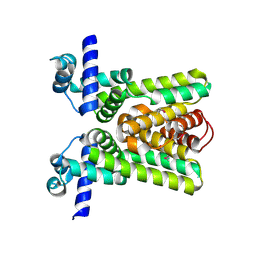 | | Crystal structure of transcriptional regulator RHA06399 from Rhodococcus sp. RHA1 | | Descriptor: | Possible transcriptional regulator, TetR family protein | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-11 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of tetR family protein.
To be Published
|
|
2QNT
 
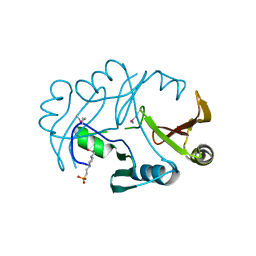 | | Crystal structure of protein of unknown function from Agrobacterium tumefaciens str. C58 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Uncharacterized protein Atu1872 | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of protein of unknown function from Agrobacterium tumefaciens str. C58.
To be Published
|
|
3DCA
 
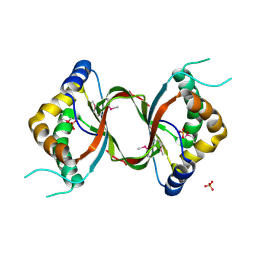 | | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target | | Descriptor: | RPA0582, SULFATE ION | | Authors: | Sledz, P, Wang, S, Chruszcz, M, Yim, V, Kudritska, M, Evdokimova, E, Turk, D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target
To be Published
|
|
3EEF
 
 | | Crystal structure of N-carbamoylsarcosine amidase from thermoplasma acidophilum | | Descriptor: | N-carbamoylsarcosine amidase related protein, ZINC ION | | Authors: | Luo, H.-B, Zheng, H, Chruszcz, M, Zimmerman, M.D, Skarina, T, Egorova, O, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and molecular modeling study of N-carbamoylsarcosine amidase Ta0454 from Thermoplasma acidophilum.
J.Struct.Biol., 169, 2010
|
|
3E6Q
 
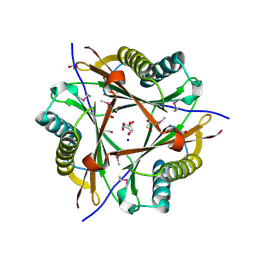 | | Putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-15 | | Release date: | 2008-08-26 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystal structure of putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa.
To be Published
|
|
3E9Q
 
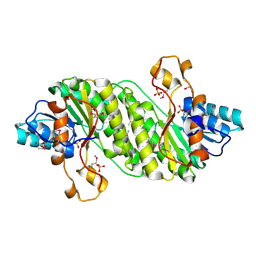 | | Crystal Structure of the Short Chain Dehydrogenase from Shigella flexneri | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-23 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Short Chain Dehydrogenase from Shigella flexneri
To be Published
|
|
4MHB
 
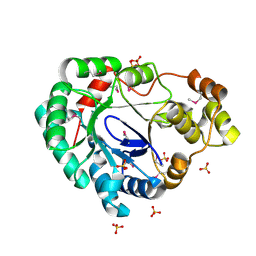 | | Structure of a putative reductase from Yersinia pestis | | Descriptor: | Putative aldo/keto reductase, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Rembert, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a putative reductase from Yersinia pestis
To be Published
|
|
3EC7
 
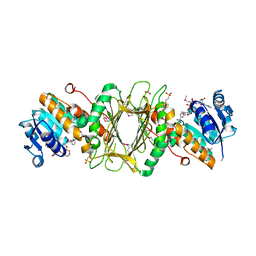 | | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETIC ACID, ... | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2
To be Published
|
|
3EFG
 
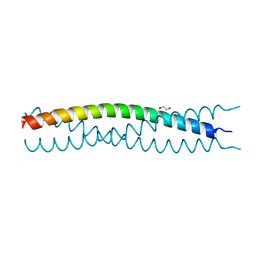 | | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, Protein slyX homolog | | Authors: | Cuff, M.E, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913
TO BE PUBLISHED
|
|
4MPH
 
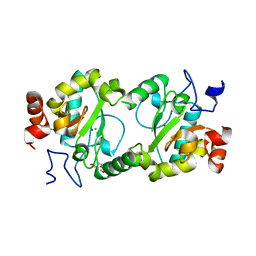 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-bound | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0301 Å) | | Cite: | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
3E7Q
 
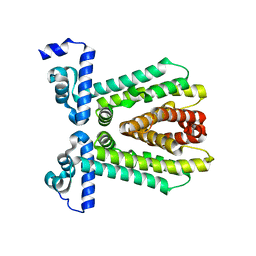 | | The crystal structure of the putative transcriptional regulator from Pseudomonas aeruginosa PAO1 | | Descriptor: | transcriptional regulator | | Authors: | Zhang, R, Skarina, T, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the putative transcriptional regulator from Pseudomonas aeruginosa PAO1
To be Published
|
|
5U08
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, ACETATE ION, CALCIUM ION, ... | | Authors: | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
3ELK
 
 | | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator TA0346 | | Authors: | Grantz Saskova, K, Chruszcz, M, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-22 | | Release date: | 2008-09-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum
To be Published
|
|
3IFS
 
 | | 2.0 Angstrom Resolution Crystal Structure of Glucose-6-phosphate Isomerase (pgi) from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Glucose-6-phosphate isomerase, LITHIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Gordon, E, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of Glucose-6-phosphate Isomerase (pgi) from Bacillus anthracis.
To be Published
|
|
3E4D
 
 | | Structural and Kinetic Study of an S-Formylglutathione Hydrolase from Agrobacterium tumefaciens | | Descriptor: | CHLORIDE ION, Esterase D, MAGNESIUM ION | | Authors: | Van Straaten, K.E, Gonzalez, C.F, Valladares, R.B, Xu, X, Savchenko, A.V, Sanders, D.A.R. | | Deposit date: | 2008-08-11 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structure of a putative S-formylglutathione hydrolase from Agrobacterium tumefaciens
Protein Sci., 18, 2009
|
|
3FG8
 
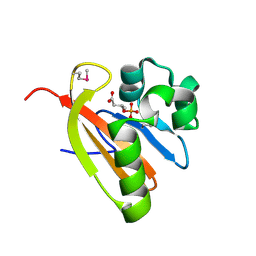 | | Crystal structure of PAS domain of RHA05790 | | Descriptor: | (3R)-3-(phosphonooxy)butanoic acid, uncharacterized protein RHA05790 | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-05 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PAS domain of RHA05790
To be Published
|
|
3EY8
 
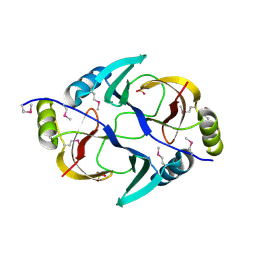 | | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1 | | Descriptor: | Biphenyl-2,3-diol 1,2-dioxygenase III-related protein, SULFATE ION | | Authors: | Harrop, S.J, Deshpande, C.N, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-20 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1
To be Published
|
|
3FBS
 
 | | The crystal structure of the oxidoreductase from Agrobacterium tumefaciens | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Oxidoreductase, ... | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the oxidoreductase from Agrobacterium tumefaciens
To be Published, 2008
|
|
3FOV
 
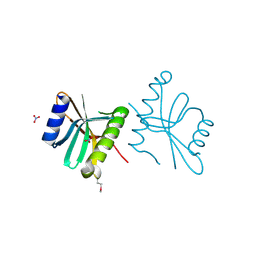 | | Crystal structure of protein RPA0323 of unknown function from Rhodopseudomonas palustris | | Descriptor: | NITRATE ION, UPF0102 protein RPA0323 | | Authors: | Osipiuk, J, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-02 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray crystal structure of protein RPA0323 of unknown function from Rhodopseudomonas palustris.
To be Published
|
|
