8WAQ
 
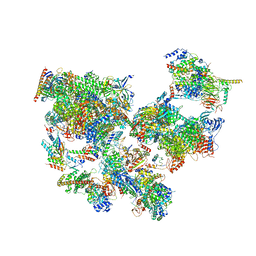 | | Structure of transcribing complex 7 (TC7), the initially transcribing complex with Pol II positioned 7nt downstream of TSS. | | 分子名称: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | 著者 | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | 登録日 | 2023-09-08 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (6.29 Å) | | 主引用文献 | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAW
 
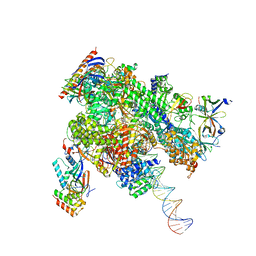 | | De novo transcribing complex 13 (TC13), the early elongation complex with Pol II positioned 13nt downstream of TSS | | 分子名称: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | 著者 | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | 登録日 | 2023-09-08 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAY
 
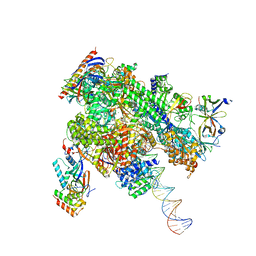 | | De novo transcribing complex 15 (TC15), the early elongation complex with Pol II positioned 15nt downstream of TSS | | 分子名称: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | 著者 | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | 登録日 | 2023-09-08 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
6L19
 
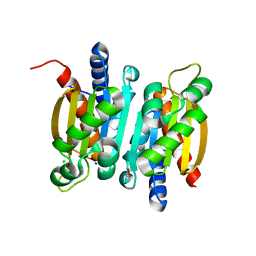 | | The crystal structure of competence or damage-inducible protein from Enterobacter asburiae | | 分子名称: | CHLORIDE ION, GLYCEROL, PncC family amidohydrolase, ... | | 著者 | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | 登録日 | 2019-09-28 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | The crystal structure of Competence or damage-inducible protein from Enterobacter asburiae
To Be Published
|
|
6L1K
 
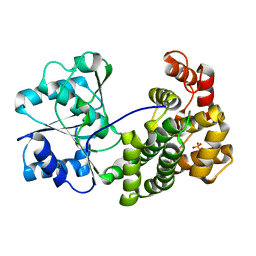 | | Crystal structure of NADH-dependent butanol dehydrogenase A from Fusobacterium nucleatum | | 分子名称: | NADH-dependent butanol dehydrogenase A, PHOSPHATE ION | | 著者 | Lan, J, Shang, F, Liu, W, Xu, Y, Chen, Y. | | 登録日 | 2019-09-29 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of NADH-dependent butanol dehydrogenase A from Fusobacterium nucleatum
To Be Published
|
|
6LQI
 
 | | Cryo-EM structure of the mouse Piezo1 isoform Piezo1.1 | | 分子名称: | Piezo-type mechanosensitive ion channel component 1 | | 著者 | Geng, J, Liu, W, Zhou, H, Zhang, T, Wang, L, Zhang, M, Shen, B, Li, X, Xiao, B. | | 登録日 | 2020-01-13 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | A Plug-and-Latch Mechanism for Gating the Mechanosensitive Piezo Channel.
Neuron, 106, 2020
|
|
8Y7R
 
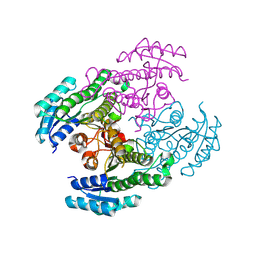 | |
8Y83
 
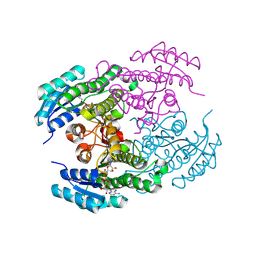 | |
7MG0
 
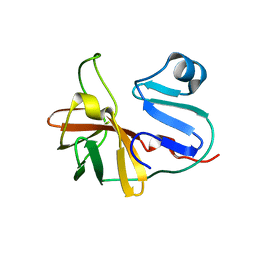 | |
6M2Z
 
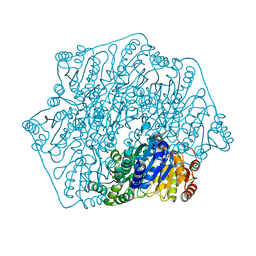 | | Crystal structure of a formolase, BFD variant M3 from Pseudomonas putida | | 分子名称: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | 著者 | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | 登録日 | 2020-03-02 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
8EFO
 
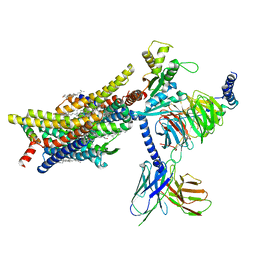 | | PZM21-bound mu-opioid receptor-Gi complex | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | 登録日 | 2022-09-08 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-30 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFQ
 
 | | DAMGO-bound mu-opioid receptor-Gi complex | | 分子名称: | DAMGO, ETHANOLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | 登録日 | 2022-09-08 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | 著者 | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | 登録日 | 2015-04-13 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
6M2Y
 
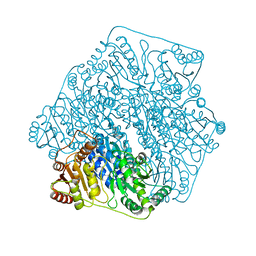 | | Crystal structure of a formolase, BFD variant M6 from Pseudomonas putida | | 分子名称: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | 著者 | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | 登録日 | 2020-03-02 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
5D8W
 
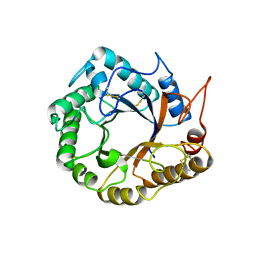 | | Structrue of a lucidum protein | | 分子名称: | Endoglucanase | | 著者 | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | 登録日 | 2015-08-18 | | 公開日 | 2016-06-29 | | 最終更新日 | 2018-05-23 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
1JYR
 
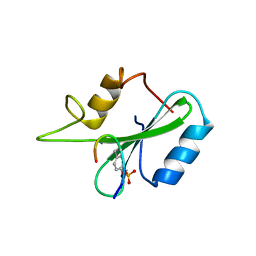 | | Xray Structure of Grb2 SH2 Domain Complexed with a Phosphorylated Peptide | | 分子名称: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, peptide: PSpYVNVQN | | 著者 | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | 登録日 | 2001-09-13 | | 公開日 | 2002-03-13 | | 最終更新日 | 2018-10-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
5D8Z
 
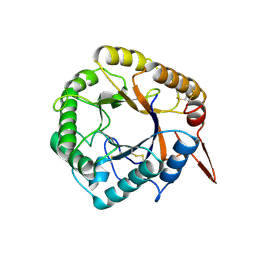 | | Structrue of a lucidum protein | | 分子名称: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, endoglucanase | | 著者 | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | 登録日 | 2015-08-18 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
1JYQ
 
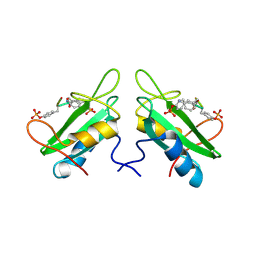 | | Xray Structure of Grb2 SH2 Domain Complexed with a Highly Affine Phospho Peptide | | 分子名称: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, mAZ-pY-(alpha Me)pY-N-NH2 peptide inhibitor | | 著者 | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | 登録日 | 2001-09-13 | | 公開日 | 2002-03-13 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
1JYU
 
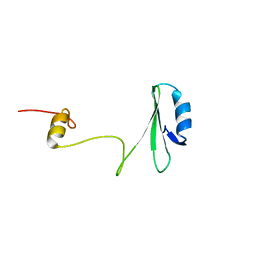 | | Xray Structure of Grb2 SH2 Domain | | 分子名称: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | 著者 | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | 登録日 | 2001-09-13 | | 公開日 | 2002-03-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
4XH7
 
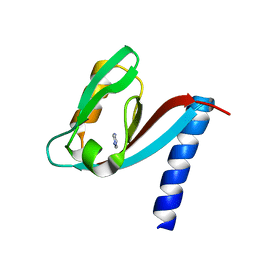 | | Crystal structure of MUPP1 PDZ4 | | 分子名称: | IMIDAZOLE, Multiple PDZ domain protein | | 著者 | Liu, Z, Zhu, H, Liu, W. | | 登録日 | 2015-01-05 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Biochemical and structural characterization of MUPP1-PDZ4 domain from Mus musculus.
Acta Biochim.Biophys.Sin., 47, 2015
|
|
7BPC
 
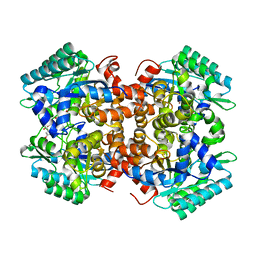 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with 2,5-DHBA | | 分子名称: | 2,3-dihydroxybenzoate decarboxylase, 2,5-dihydroxybenzoic acid, ZINC ION | | 著者 | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | 登録日 | 2020-03-22 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
7BP1
 
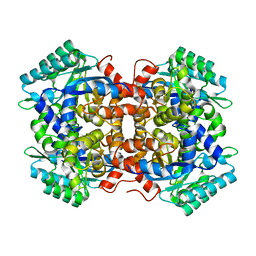 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with Catechol | | 分子名称: | 2,3-dihydroxybenzoate decarboxylase, CATECHOL, ZINC ION | | 著者 | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | 登録日 | 2020-03-21 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
6W8T
 
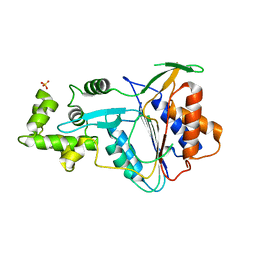 | | Crystal structure of metacaspase 4 from Arabidopsis (microcrystals treated with calcium) | | 分子名称: | Metacaspase-4, SULFATE ION | | 著者 | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | 登録日 | 2020-03-21 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8R
 
 | | Crystal structure of metacaspase 4 C139A from Arabidopsis | | 分子名称: | Metacaspase-4, SULFATE ION | | 著者 | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | 登録日 | 2020-03-21 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8S
 
 | | Crystal structure of metacaspase 4 from Arabidopsis | | 分子名称: | Metacaspase-4, SULFATE ION | | 著者 | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | 登録日 | 2020-03-21 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.484 Å) | | 主引用文献 | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
