1SZU
 
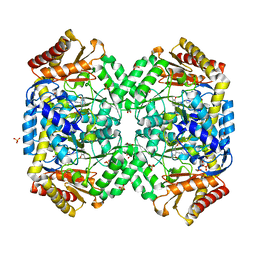 | | The structure of gamma-aminobutyrate aminotransferase mutant: V241A | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | 著者 | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | 登録日 | 2004-04-06 | | 公開日 | 2005-03-01 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1SZS
 
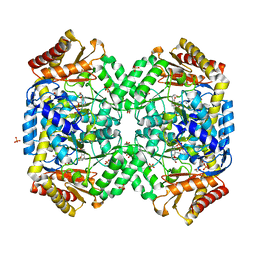 | | The structure of gamma-aminobutyrate aminotransferase mutant: I50Q | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | 著者 | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | 登録日 | 2004-04-06 | | 公開日 | 2005-03-01 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1SZK
 
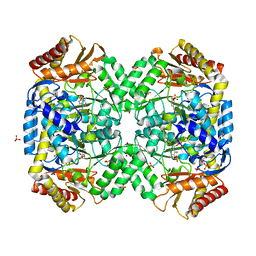 | | The structure of gamma-aminobutyrate aminotransferase mutant: E211S | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | 著者 | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Fisher, A.J, Toney, M.D. | | 登録日 | 2004-04-05 | | 公開日 | 2005-03-01 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
7MSJ
 
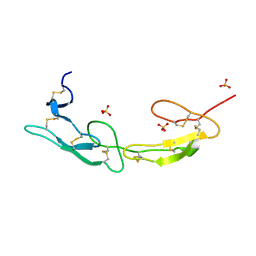 | | The crystal structure of mouse HVEM | | 分子名称: | SULFATE ION, Tumor necrosis factor receptor superfamily member 14 | | 著者 | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | 登録日 | 2021-05-11 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7MSG
 
 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | 著者 | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | 登録日 | 2021-05-11 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7TDO
 
 | | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | 著者 | Liu, W, Schoonen, M, Wang, T, McSweeney, S, Liu, Q. | | 登録日 | 2022-01-02 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state.
Commun Biol, 5, 2022
|
|
5DBG
 
 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NAD+ | | 分子名称: | Iridoid synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Liu, W.D, Hu, Y.M, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | 登録日 | 2015-08-21 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1XD6
 
 | | Crystal structures of novel monomeric monocot mannose-binding lectins from Gastrodia elata | | 分子名称: | SULFATE ION, gastrodianin-4 | | 著者 | Liu, W, Yang, N, Wang, M, Huang, R.H, Hu, Z, Wang, D.C. | | 登録日 | 2004-09-04 | | 公開日 | 2005-01-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Mechanism Governing the Quaternary Organization of Monocot Mannose-binding Lectin Revealed by the Novel Monomeric Structure of an Orchid Lectin
J.Biol.Chem., 280, 2005
|
|
1ZOD
 
 | |
1ZOB
 
 | |
1IR5
 
 | |
2VL6
 
 | | STRUCTURAL ANALYSIS OF THE SULFOLOBUS SOLFATARICUS MCM PROTEIN N- TERMINAL DOMAIN | | 分子名称: | MINICHROMOSOME MAINTENANCE PROTEIN MCM, ZINC ION | | 著者 | Liu, W, Pucci, B, Rossi, M, Pisani, F.M, Ladenstein, R. | | 登録日 | 2008-01-08 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Analysis of the Sulfolobus Solfataricus Mcm Protein N-Terminal Domain.
Nucleic Acids Res., 36, 2008
|
|
3WB9
 
 | | Crystal Structures of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum | | 分子名称: | Diaminopimelate dehydrogenase, GLYCEROL | | 著者 | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | 登録日 | 2013-05-14 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural and mutational studies on the unusual substrate specificity of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum.
Chembiochem, 15, 2014
|
|
4OLT
 
 | | Chitosanase complex structure | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Chitosanase, GLYCEROL | | 著者 | Liu, W.Z, Lyu, Q.Q, Han, B.Q. | | 登録日 | 2014-01-25 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural insights into the substrate-binding mechanism for a novel chitosanase.
Biochem.J., 461, 2014
|
|
3WBB
 
 | | Crystal Structures of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum | | 分子名称: | Diaminopimelate dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | 登録日 | 2013-05-14 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural and mutational studies on the unusual substrate specificity of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum.
Chembiochem, 15, 2014
|
|
5L19
 
 | |
6LNZ
 
 | |
5I78
 
 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | 著者 | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | 登録日 | 2016-02-17 | | 公開日 | 2016-12-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
5I79
 
 | | Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | 著者 | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | 登録日 | 2016-02-17 | | 公開日 | 2016-12-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
3N9T
 
 | | Cryatal structure of Hydroxyquinol 1,2-dioxygenase from Pseudomonas putida DLL-E4 | | 分子名称: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, CITRATE ANION, FE (III) ION, ... | | 著者 | Liu, W, Shen, W, Fang, P, Li, J, Cui, Z. | | 登録日 | 2010-05-31 | | 公開日 | 2010-08-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Cryatal structure of Hydroxyquinol 1,2-dioxygenase from Pseudomonas putida DLL-E4
To be Published
|
|
6FD2
 
 | | Radical SAM 1,2-diol dehydratase AprD4 in complex with its substrate paromamine | | 分子名称: | 5'-DEOXYADENOSINE, IRON/SULFUR CLUSTER, METHIONINE, ... | | 著者 | Liu, W.Q, Amara, P, Mouesca, J.M, Ji, X, Renoux, O, Martin, L, Zhang, C, Zhang, Q, Nicolet, Y. | | 登録日 | 2017-12-21 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | 1,2-Diol Dehydration by the Radical SAM Enzyme AprD4: A Matter of Proton Circulation and Substrate Flexibility.
J. Am. Chem. Soc., 140, 2018
|
|
4RSU
 
 | | Crystal structure of the light and hvem complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | 著者 | Liu, W, Ramagoal, U.A, Himmel, D, Bonanno, J.B, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-11-11 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
5L36
 
 | |
3D6V
 
 | | Crystal structure of 4-(trifluoromethyldiazirinyl)phenylalanyl-tRNA synthetase | | 分子名称: | 4-(2,2,2-TRIFLUOROETHYL)-L-PHENYLALANINE, BETA-MERCAPTOETHANOL, Tyrosyl-tRNA synthetase | | 著者 | Liu, W, Tippmann, E, Mack, A.V, Schultz, P.G. | | 登録日 | 2008-05-20 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A genetically encoded diazirine photocrosslinker in Escherichia coli
ChemBioChem, 8, 2007
|
|
5Z9T
 
 | | a new PL6 alginate lyase complex with trisaccharide | | 分子名称: | GLYCEROL, MALONATE ION, SODIUM ION, ... | | 著者 | Liu, W.Z, Lyu, Q.Q, Zhang, K.K, Li, Z.J. | | 登録日 | 2018-02-05 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into a novel Ca2+-independent PL-6 alginate lyase from Vibrio OU02 identify the possible subsites responsible for product distribution.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
