1PUZ
 
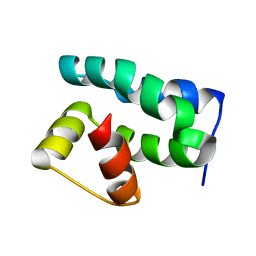 | | Solution NMR Structure of Protein NMA1147 from Neisseria meningitidis. Northeast Structural Genomics Consortium Target MR19 | | Descriptor: | conserved hypothetical protein | | Authors: | Liu, G, Xu, D, Sukumaran, D.K, Chiang, Y, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-06-25 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the hypothetical protein NMA1147 from Neisseria meningitidis reveals a distinct 5-helix bundle.
Proteins, 55, 2004
|
|
1S04
 
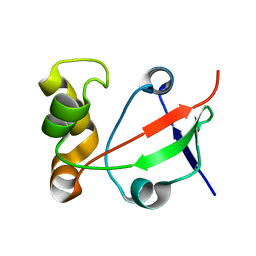 | | Solution NMR Structure of Protein PF0455 from Pyrococcus furiosus. Northeast Structural Genomics Consortium Target PfR13 | | Descriptor: | hypothetical protein PF0455 | | Authors: | liu, G, Xiao, R, Sukumaran, D.K, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-29 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of The Hypothetical Protein PF0455 From Pyrococcus furiosus: Northeast Structural Genomics Consortium Target PfR13
To be Published
|
|
2JWY
 
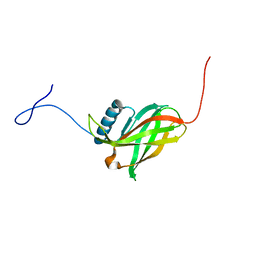 | | Solution NMR structure of uncharacterized lipoprotein yajI from Escherichia coli. Northeast Structural Genomics target ER540 | | Descriptor: | Uncharacterized lipoprotein yajI | | Authors: | Liu, G, Xiao, R, Chen, C, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of uncharacterized lipoprotein yajI from Escherichia coli.
To be Published
|
|
1TUZ
 
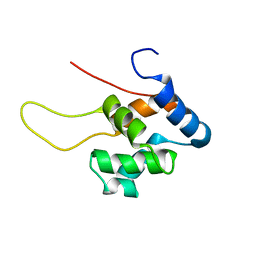 | | NMR Structure of the Diacylglycerol kinase alpha, NESGC target HR532 | | Descriptor: | diacylglycerol kinase alpha | | Authors: | Liu, G, Shao, Y, Xiao, R, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Diacylglycerol kinase alpha, NESGC target HR532
To be Published
|
|
2KCV
 
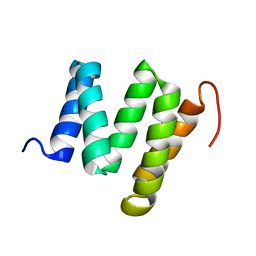 | | Solution nmr structure of tetratricopeptide repeat domain protein sru_0103 from salinibacter ruber, northeast structural genomics consortium (nesg) target srr115c | | Descriptor: | Tetratricopeptide repeat domain protein | | Authors: | Liu, G, Rossi, P, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution nmr structure of tetratricopeptide repeat domain protein sru_0103 from salinibacter ruber, northeast structural genomics consortium (nesg) target srr115c
To be Published
|
|
2JXT
 
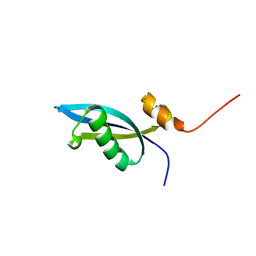 | | Solution structure of 50S ribosomal protein LX from Methanobacterium thermoautotrophicum. Northeast Structural Genomics Consortium (NESG) target TR80 | | Descriptor: | 50S ribosomal protein LX | | Authors: | Liu, G, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 50S ribosomal protein LX from Methanobacterium thermoautotrophicum.
To be Published
|
|
2KCP
 
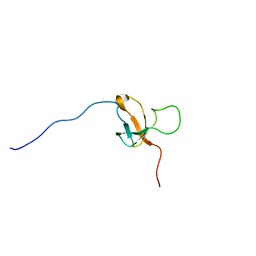 | | SOLUTION STRUCTURE OF 30S RIBOSOMAL PROTEIN S8E; FROM Methanothermobacter thermautotrophicus, NORTHEASTSTRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET Tr71d | | Descriptor: | 30S ribosomal protein S8e | | Authors: | Liu, G, Rossi, P, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-24 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF 30S RIBOSOMAL PROTEIN S8E FROM Methanothermobacter thermautotrophicus, NORTHEASTSTRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET
To be Published
|
|
1PH0
 
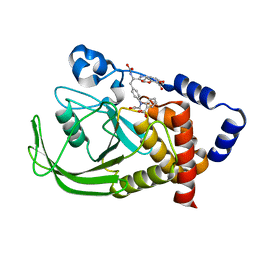 | | Non-carboxylic Acid-Containing Inhibitor of PTP1B Targeting the Second Phosphotyrosine Site | | Descriptor: | 2-{4-[2-(S)-ALLYLOXYCARBONYLAMINO-3-{4-[(2-CARBOXY-PHENYL)-OXALYL-AMINO]-PHENYL}-PROPIONYLAMINO]-BUTOXY}-6-HYDROXY-BENZ OIC ACID METHYL ESTER, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Xin, Z, Liang, H, Abad-Zapatero, C, Hajduk, P, Janowick, D, Szczepankiewicz, B, Pei, Z, Hutchins, C.W, Ballaron, S.J. | | Deposit date: | 2003-05-29 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Protein Tyrosine Phosphatase 1B Inhibitors: Targeting the Second Phosphotyrosine Binding Site with Non-Carboxylic Acid-Containing Ligands.
J.Med.Chem., 46, 2003
|
|
2KCY
 
 | | SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S8E FROM Methanothermobacter thermautotrophicus, NORTHEASTSTRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET TR71D | | Descriptor: | 30S ribosomal protein S8e | | Authors: | Liu, G, Rossi, P, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B.C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-30 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S8E FROM Methanothermobacter thermautotrophicus, NORTHEASTSTRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET TR71D
To be Published
|
|
2KK1
 
 | | Solution structure of C-terminal Domain of Tyrosine-protein kinase ABL2 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) target HR5537A | | Descriptor: | Tyrosine-protein kinase ABL2 | | Authors: | Liu, G, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-14 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of F-actin-binding domain of Arg/Abl2 from Homo sapiens.
Proteins, 78, 2010
|
|
2KK0
 
 | | Solution structure of dead ringer-like protein 1 (at-rich interactive domain-containing protein 3a) from homo sapiens, northeast structural genomics consortium (NESG) target hr4394c | | Descriptor: | AT-rich interactive domain-containing protein 3A | | Authors: | Liu, G, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-14 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ARID domain of human AT-rich interactive domain-containing protein 3A: a human cancer protein interaction network target.
Proteins, 78, 2010
|
|
8H0L
 
 | | Sulfur binding domain of Hga complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*(PST)P*TP*CP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*GP*AP*AP*CP*TP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Liu, G, He, X, Hu, W, Yang, B, Xiao, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a promiscuous DNA sulfur binding domain and application in site-directed RNA base editing.
Nucleic Acids Res., 51, 2023
|
|
7KBQ
 
 | |
7CNT
 
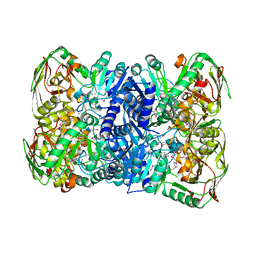 | | Structure of 2,5-dihydroxypridine Dioxygenase from Pseudomonas putida KT2440 in complex with product N-formylmaleamic acid formed via in crystallo reaction with 2,5-dihydroxypridine | | Descriptor: | 2,5-dihydroxypyridine 5,6-dioxygenase, FE (II) ION | | Authors: | Liu, G.Q, Tang, H.Z. | | Deposit date: | 2020-08-03 | | Release date: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | 2,5-dihydroxypridine Dioxygenase in complex with 2,5-dihydroxypridine and product N-formylmaleamic acid
Nat Commun, 2020
|
|
7CUP
 
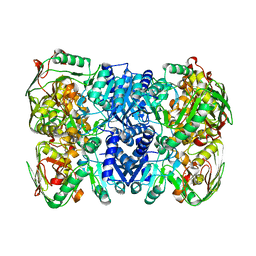 | | Structure of 2,5-dihydroxypridine Dioxygenase from Pseudomonas putida KT2440 | | Descriptor: | 2,5-dihydroxypyridine 5,6-dioxygenase, FE (III) ION | | Authors: | Liu, G.Q, Tang, H.Z. | | Deposit date: | 2020-08-23 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2,5-dihydroxypridine Dioxygenase in complex with 2,5-dihydroxypridine and product N-formylmaleamic acid
Nat Commun, 2020
|
|
7CN3
 
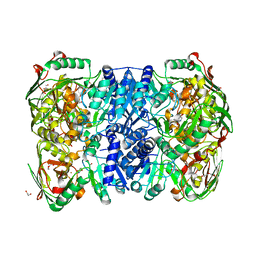 | | 2,5-dihydroxypridine Dioxygenase in complex with 2,5-dihydroxypridine and product N-formylmaleamic acid | | Descriptor: | (~{Z})-4-formamido-4-oxidanylidene-but-2-enoic acid, 1,2-ETHANEDIOL, 2,5-dihydroxypyridine 5,6-dioxygenase, ... | | Authors: | Liu, G.Q, Tang, H.Z. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2,5-dihydroxypridine Dioxygenase in complex with 2,5-dihydroxypridine and product N-formylmaleamic acid
Nat Commun, 2020
|
|
8JIY
 
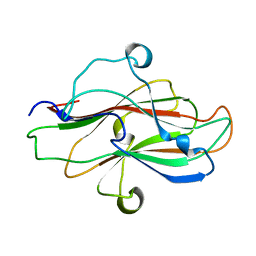 | | A carbohydrate binding domain of a putative chondroitinase | | Descriptor: | DUF4955 domain-containing protein | | Authors: | Liu, G.C, Chang, Y.G. | | Deposit date: | 2023-05-29 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and structural characterization of a novel chondroitin sulfate-specific carbohydrate-binding module: The first member of a new family, CBM100.
Int.J.Biol.Macromol., 255, 2024
|
|
4M1K
 
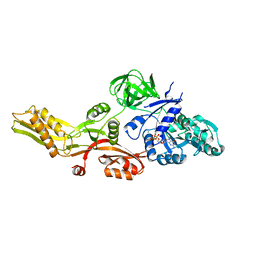 | |
5GAJ
 
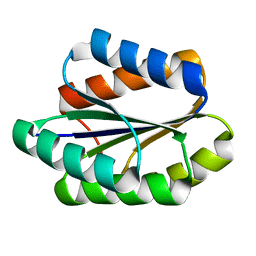 | | Solution NMR structure of De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258 | | Descriptor: | DE NOVO DESIGNED PROTEIN OR258 | | Authors: | Liu, G, Castelllanos, J, Koga, R, Koga, N, Xiao, R, Pederson, K, Janjua, H, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258
To Be Published
|
|
4MYU
 
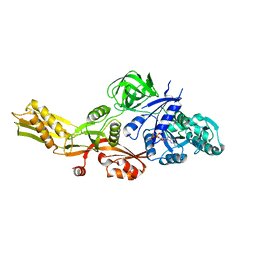 | |
4MYT
 
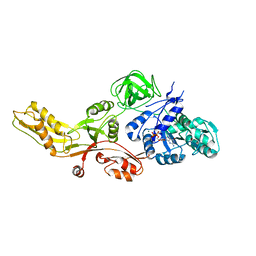 | |
2K5E
 
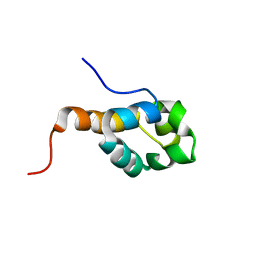 | | SOLUTION STRUCTURE OF PUTATIVE UNCHARACTERIZED PROTEIN GSU1278 FROM METHANOCALDOCOCCUS JANNASCHII, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET GsR195 | | Descriptor: | uncharacterized protein | | Authors: | Liu, G, Zhao, L, Ciccosanti, C, Jiang, M, Xiao, R, Swapna, G, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF PUTATIVE UNCHARACTERIZED PROTEIN GSU1278 FROM METHANOCALDOCOCCUS JANNASCHII, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET GsR195
To be Published
|
|
8WAB
 
 | |
2LVB
 
 | | Solution NMR Structure DE NOVO DESIGNED PFK fold PROTEIN, Northeast Structural Genomics Consortium (NESG) Target OR250 | | Descriptor: | DE NOVO DESIGNED PFK fold PROTEIN | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-30 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2MBL
 
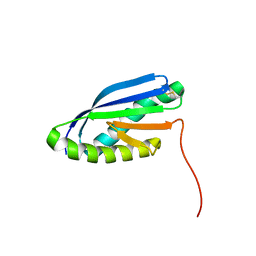 | | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33 | | Descriptor: | Top7 Fold Protein Top7m13 | | Authors: | Liu, G, Zanghellini, A.L, Chan, K, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-08-02 | | Release date: | 2013-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33
To be Published
|
|
