5AER
 
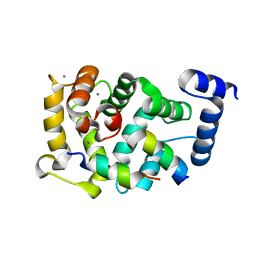 | | Neuronal calcium sensor-1 (NCS-1)from Rattus norvegicus complex with D2 dopamine receptor peptide from Homo sapiens | | 分子名称: | CALCIUM ION, D(2) DOPAMINE RECEPTOR, NEURONAL CALCIUM SENSOR 1, ... | | 著者 | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | 登録日 | 2015-01-08 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
5AEQ
 
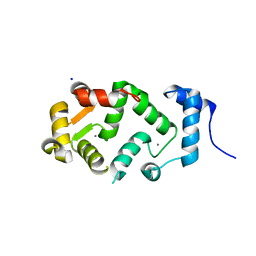 | | Neuronal calcium sensor (NCS-1)from Rattus norvegicus | | 分子名称: | CALCIUM ION, NEURONAL CALCIUM SENSOR 1, SODIUM ION | | 著者 | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | 登録日 | 2015-01-08 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
5DCK
 
 | |
4V46
 
 | | Crystal structure of the BAFF-BAFF-R complex | | 分子名称: | MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B, Tumor necrosis factor receptor superfamily member 13C | | 著者 | Kim, H.M, Yu, K.S, Lee, M.E, Shin, D.R, Kim, Y.S, Paik, S.G, Yoo, O.J, Lee, H, Lee, J.-O. | | 登録日 | 2003-03-23 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation
NAT.STRUCT.BIOL., 10, 2003
|
|
4UW2
 
 | | Crystal structure of Csm1 in T.onnurineus | | 分子名称: | CSM1 | | 著者 | Jung, T.Y, An, Y, Park, K.H, Lee, M.H, Oh, B.H, Woo, E.J. | | 登録日 | 2014-08-08 | | 公開日 | 2015-03-25 | | 最終更新日 | 2015-09-23 | | 実験手法 | X-RAY DIFFRACTION (2.632 Å) | | 主引用文献 | Crystal Structure of the Csm1 Subunit of the Csm Complex and its Single-Stranded DNA-Specific Nuclease Activity.
Structure, 23, 2015
|
|
7MG0
 
 | |
2Y2C
 
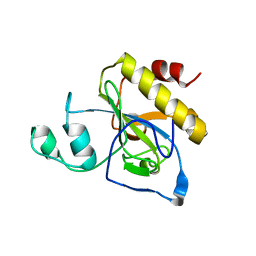 | | crystal structure of AmpD Apoenzyme | | 分子名称: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD | | 著者 | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | 登録日 | 2010-12-14 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y28
 
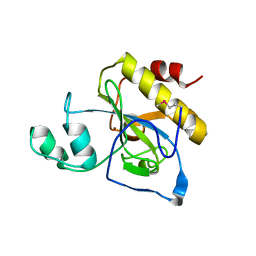 | | crystal structure of Se-Met AmpD derivative | | 分子名称: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | 著者 | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | 登録日 | 2010-12-14 | | 公開日 | 2011-07-20 | | 最終更新日 | 2011-12-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2E
 
 | | crystal structure of AmpD grown at pH 5.5 | | 分子名称: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | 著者 | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | 登録日 | 2010-12-14 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2D
 
 | | crystal structure of AmpD holoenzyme | | 分子名称: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | 著者 | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | 登録日 | 2010-12-14 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2B
 
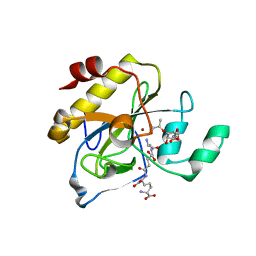 | | crystal structure of AmpD in complex with reaction products | | 分子名称: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | 著者 | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | 登録日 | 2010-12-14 | | 公開日 | 2011-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
5KUC
 
 | | Crystal structure of trypsin activated Cry6Aa | | 分子名称: | Pesticidal crystal protein Cry6Aa | | 著者 | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Hey, T, Chikwana, V.M, Narva, K.E. | | 登録日 | 2016-07-13 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
5KUD
 
 | | Crystal structure of full length Cry6Aa | | 分子名称: | Pesticidal crystal protein Cry6Aa | | 著者 | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Chikwana, V.M, Hey, T, Narva, K. | | 登録日 | 2016-07-13 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
4OCA
 
 | | Cryatal structure of ArnB K188A complexted with PLP and UDP-Ara4N | | 分子名称: | (2R,3R,4S,5S)-3,4-dihydroxy-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]tetrahydro-2H-pyr an-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase | | 著者 | Sousa, M.C, Lee, M. | | 登録日 | 2014-01-08 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis for Substrate Specificity in ArnB. A Key Enzyme in the Polymyxin Resistance Pathway of Gram-Negative Bacteria.
Biochemistry, 53, 2014
|
|
5KW2
 
 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | 分子名称: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | 著者 | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | 登録日 | 2016-07-15 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
1VZK
 
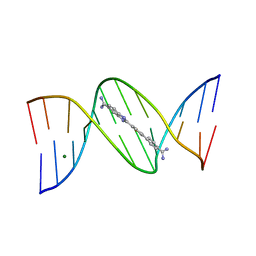 | | A Thiophene Based Diamidine Forms a "Super" AT Binding Minor Groove Agent | | 分子名称: | 2-(5-{4-[AMINO(IMINO)METHYL]PHENYL}-2-THIENYL)-1H-BENZIMIDAZOLE-6- CARBOXIMIDAMIDE DIHYDROCHLORIDE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP *CP*G)-3', MAGNESIUM ION | | 著者 | Mallena, S, Lee, M.P.H, Bailly, C, Neidle, S, Kumar, A, Boykin, D.W, Wilson, W.D. | | 登録日 | 2004-05-20 | | 公開日 | 2004-10-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Thiophene-Based Diamidine Forms a "Super" at Binding Minor Groove Agent
J.Am.Chem.Soc., 142, 2004
|
|
6DLN
 
 | |
6KTA
 
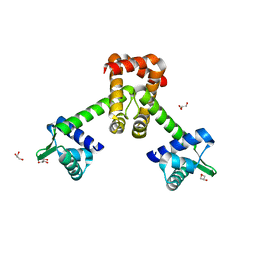 | | Crystal structure of B. halodurans MntR in apo form | | 分子名称: | GLYCEROL, HTH-type transcriptional regulator MntR | | 著者 | Lee, J.Y, Lee, M.Y. | | 登録日 | 2019-08-26 | | 公開日 | 2019-12-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural analysis of the manganese transport regulator MntR from Bacillus halodurans in apo and manganese bound forms.
Plos One, 14, 2019
|
|
7CV0
 
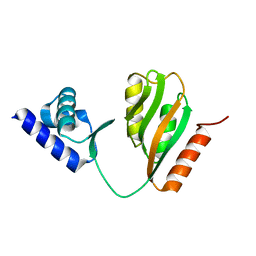 | | Crystal structure of B. halodurans NiaR in apo form | | 分子名称: | Transcriptional regulator NiaR, ZINC ION | | 著者 | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | 登録日 | 2020-08-25 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
7QAJ
 
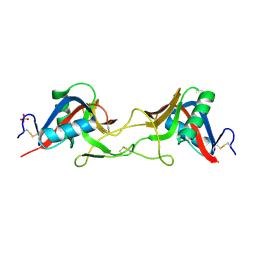 | | ZK002 with Anti-angiogenic and Anti-inflamamtory Properties | | 分子名称: | SULFATE ION, Snaclec clone 2100755 alpha, Snaclec clone 2100755 beta | | 著者 | Wong, W.Y, Chan, B.D, Muk Lan Lee, M, Dai, X, Tsim, K.W.K, Hsiao, W.L.W, Li, M, Li, X.Y, Tai, W.C.S. | | 登録日 | 2021-11-17 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Isolation and characterization of ZK002, a novel dual function snake venom protein from Deinagkistrodon acutus with anti-angiogenic and anti-inflammatory properties.
Front Pharmacol, 14, 2023
|
|
6UE0
 
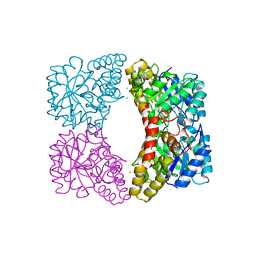 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | 分子名称: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | 著者 | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | 登録日 | 2019-09-20 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.892 Å) | | 主引用文献 | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
7CV2
 
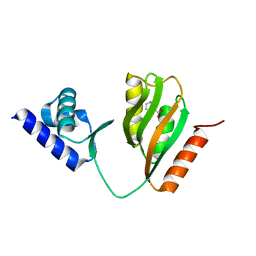 | | Crystal structure of B. halodurans NiaR in niacin-bound form | | 分子名称: | NICOTINIC ACID, Transcriptional regulator NiaR, ZINC ION | | 著者 | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | 登録日 | 2020-08-25 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
6Z3C
 
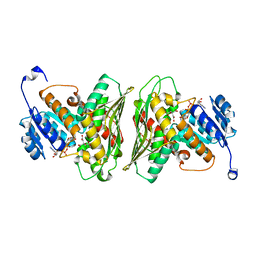 | | High resolution structure of RgNanOx | | 分子名称: | CITRATE ANION, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Naismith, J.H, Lee, M.O. | | 登録日 | 2020-05-19 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
6XFM
 
 | | Molecular structure of the core of amyloid-like fibrils formed by residues 111-214 of FUS | | 分子名称: | RNA-binding protein FUS | | 著者 | Tycko, R, Lee, M, Ghosh, U, Thurber, K, Kato, M. | | 登録日 | 2020-06-15 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Molecular structure and interactions within amyloid-like fibrils formed by a low-complexity protein sequence from FUS.
Nat Commun, 11, 2020
|
|
6LCI
 
 | | Solution structure of mdaA-1 domain | | 分子名称: | mdaA-1 | | 著者 | Nguyen, T.A, Le, S, Lee, M, Fan, J.S, Yang, D, Yan, J, Jedd, G. | | 登録日 | 2019-11-19 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Fungal Wound Healing through Instantaneous Protoplasmic Gelation.
Curr.Biol., 31, 2021
|
|
