8S7X
 
 | | Methyl-coenzyme M reductase activation complex without the A2 component | | Descriptor: | 1-THIOETHANESULFONIC ACID, Coenzyme B, DUF2098 domain-containing protein, ... | | Authors: | Ramirez-Amador, F, Paul, S, Kumar, A, Schuller, J.M. | | Deposit date: | 2024-03-04 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Methyl-coenzyme M reductase activation complex without the A2 component
To Be Published
|
|
6IGP
 
 | | Crystal structure of S9 peptidase (inactive state)from Deinococcus radiodurans R1 in P212121 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IGQ
 
 | | Crystal structure of inactive state of S9 peptidase from Deinococcus radiodurans R1 (PMSF treated) | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL, ... | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
1RL4
 
 | | Plasmodium falciparum peptide deformylase complex with inhibitor | | Descriptor: | (2R)-2-{[FORMYL(HYDROXY)AMINO]METHYL}HEXANOIC ACID, 2-{N'-[2-(5-AMINO-1-PHENYLCARBAMOYL-PENTYLCARBAMOYL)-HEXYL]-HYDRAZINOMETHYL}-HEXANOIC ACID(5-AMINO-1-PHENYLCARBAMOYL-PENTYL)-AMIDE, COBALT (II) ION, ... | | Authors: | Robien, M.A, Nguyen, K.T, Kumar, A, Hirsh, I, Turley, S, Pei, D, Hol, W.G.J. | | Deposit date: | 2003-11-24 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | An improved crystal form of Plasmodium falciparum peptide deformylase.
Protein Sci., 13, 2004
|
|
6IX1
 
 | |
7FDP
 
 | | Crystal structure of PirB insecticidal protein from Photorhabdus akhurstii | | Descriptor: | Insecticidal protein | | Authors: | Prashar, A, Kinkar, O, Kumar, A, Hire, R.S, Makde, R.D. | | Deposit date: | 2021-07-17 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of PirA and PirB toxins from Photorhabdus akhurstii subsp. akhurstii K-1
Insect Biochem.Mol.Biol., 162, 2023
|
|
5YZO
 
 | | Crystal structure of S9 peptidase mutant (S514A) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative, DIMETHYL SULFOXIDE, ... | | Authors: | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
1RQC
 
 | | Crystals of peptide deformylase from Plasmodium falciparum with ten subunits per asymmetric unit reveal critical characteristics of the active site for drug design | | Descriptor: | COBALT (II) ION, formylmethionine deformylase | | Authors: | Robien, M.A, Nguyen, K.T, Kumar, A, Hirsh, I, Turley, S, Pei, D, Hol, W.G. | | Deposit date: | 2003-12-04 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An improved crystal form of Plasmodium falciparum peptide deformylase
Protein Sci., 13, 2004
|
|
8HMR
 
 | | Crystal Structure of PKM2 mutant L144P | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Upadhyay, S, Kumar, A, Patel, A.K. | | Deposit date: | 2022-12-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and mechanistic insights into cancer patient-derived mutations in Pyruvate Kinase muscle isoform 2
To Be Published
|
|
5XEV
 
 | | Crystal Structure of a novel Xaa-Pro dipeptidase from Deinococcus radiodurans | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Are, V.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-04-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a novel prolidase from Deinococcus radiodurans identifies new subfamily of bacterial prolidases.
Proteins, 85, 2017
|
|
6M1C
 
 | |
7FCN
 
 | | Crystal strcture of PirA insecticidal protein from Photorhabdus akhurstii | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Prashar, A, Kumar, A, Kinkar, O, Hire, R.S, Makde, R.D. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of PirA and PirB toxins from Photorhabdus akhurstii subsp. akhurstii K-1
Insect Biochem.Mol.Biol., 162, 2023
|
|
5GIU
 
 | | Crystal structure of Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site | | Descriptor: | PHOSPHATE ION, Proline dipeptidase, SODIUM ION | | Authors: | Are, V.N, Kumar, A, Singh, R, Ghosh, B, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-25 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site
To Be Published
|
|
8TNT
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with antibodies F-2-1 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 heavy chain, 769C2 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
1BPE
 
 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA; EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA POLYMERASE BETA | | Authors: | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-04-12 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
2NYQ
 
 | |
3P20
 
 | | Crystal structure of vanadate bound subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-10-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The transition-like state and Pi entrance into the catalytic a subunit of the biological engine A-ATP synthase.
J.Mol.Biol., 408, 2011
|
|
2QF8
 
 | | Crystal structure of the complex of Buffalo Secretory Glycoprotein with tetrasaccharide at 2.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Singh, A.K, Jain, R, Sinha, M, Kumar, A, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the complex of Buffalo Secretory Glycoprotein with Tetrasaccharide at 2.8A resolution
To be Published
|
|
6I1D
 
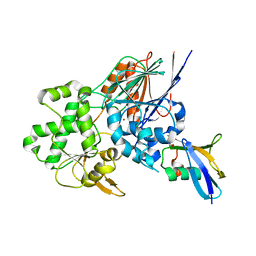 | | Structure of the Ysh1-Mpe1 nuclease complex from S.cerevisiae | | Descriptor: | Endoribonuclease YSH1, GLYCEROL, Protein MPE1, ... | | Authors: | Hill, C.H, Boreikaite, V, Kumar, A, Casanal, A, Kubik, P, Degliesposti, G, Maslen, S, Mariani, A, von Loeffelholz, O, Girbig, M, Skehel, M, Passmore, L.A. | | Deposit date: | 2018-10-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Activation of the Endonuclease that Defines mRNA 3' Ends Requires Incorporation into an 8-Subunit Core Cleavage and Polyadenylation Factor Complex.
Mol.Cell, 73, 2019
|
|
5GIQ
 
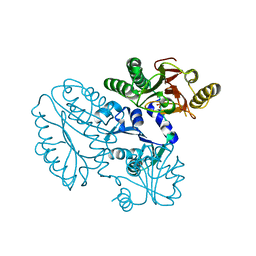 | | Xaa-Pro peptidase from Deinococcus radiodurans, Zinc bound | | Descriptor: | PHOSPHATE ION, Proline dipeptidase, ZINC ION | | Authors: | Are, V.N, Singh, R, Kumar, A, Ghosh, B, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases.
Proteins, 2018
|
|
9BD8
 
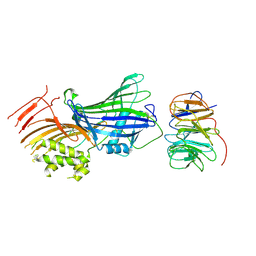 | | ApoB 100 beta barrel bound to LDLR beta propeller | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Apolipoprotein B-100, Low-density lipoprotein receptor | | Authors: | Dearborn, A.D, Reimund, M, Graziano, G, Lei, H, Kumar, A, Neufeld, E.B, Remaley, A.T, Marcotrigiano, J. | | Deposit date: | 2024-04-11 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of apolipoprotein B100 bound to the low-density lipoprotein receptor.
Nature, 638, 2025
|
|
9BDE
 
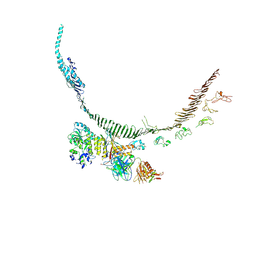 | | Middle Region of Apolipoprotein B 100 bound to Low Density Lipoprotein Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ApoB100 nanobody 4, Apolipoprotein B 100, ... | | Authors: | Dearborn, A.D, Reimund, M, Graziano, G, Lei, H, Kumar, A, Neufeld, E.B, Remaley, A.T, Marcotrigiano, J. | | Deposit date: | 2024-04-11 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structure of apolipoprotein B100 bound to the low-density lipoprotein receptor.
Nature, 638, 2025
|
|
9BD1
 
 | | beta/alpha1 region of ApoB 100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Apolipoprotein B-100 | | Authors: | Dearborn, A.D, Reimund, M, Graziano, G, Lei, H, Kumar, A, Neufeld, E.B, Remaley, A.T, Marcotrigiano, J. | | Deposit date: | 2024-04-10 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of apolipoprotein B100 bound to the low-density lipoprotein receptor.
Nature, 638, 2025
|
|
9BDT
 
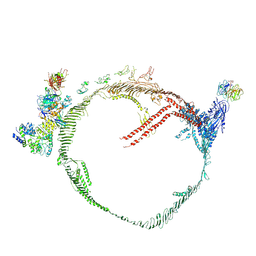 | | Apolipoprotein B 100 bound to LDL receptor and legobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ApoB100 nanobody 4, ... | | Authors: | Dearborn, A.D, Reimund, M, Graziano, G, Lei, H, Kumar, A, Neufeld, E.B, Remaley, A.T, Marcotrigiano, J. | | Deposit date: | 2024-04-12 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of apolipoprotein B100 bound to the low-density lipoprotein receptor.
Nature, 638, 2025
|
|
8HMS
 
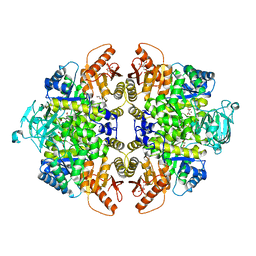 | | Crystal Structure of PKM2 mutant C474S | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, ... | | Authors: | Upadhyay, S, Kumar, A, Patel, A.K. | | Deposit date: | 2022-12-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic insights into cancer patient-derived mutations in Pyruvate Kinase muscle isoform 2
To Be Published
|
|
