1AGC
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYQL-7Q MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYQL - 7Q MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1GP0
 
 | | Human IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, SULFATE ION | | Authors: | Brown, J, Esnouf, R.M, Jones, M.A, Linnell, J, Harlos, K, Hassan, A.B, Jones, E.Y. | | Deposit date: | 2001-10-29 | | Release date: | 2002-02-28 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a Functional Igf2R Fragment Determined from the Anomalous Scattering of Sulfur
Embo J., 21, 2002
|
|
1GP3
 
 | | Human IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Brown, J, Esnouf, R.M, Jones, M.A, Linnell, J, Harlos, K, Hassan, A.B, Jones, E.Y. | | Deposit date: | 2001-10-29 | | Release date: | 2002-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a Functional Igf2R Fragment Determined from the Anomalous Scattering of Sulfur
Embo J., 21, 2002
|
|
1GZP
 
 | | CD1b in complex with GM2 ganglioside | | Descriptor: | B2-MICROGLOBULIN, DOCOSANE, DODECANE, ... | | Authors: | Gadola, S.D, Zaccai, N.R, Harlos, K, Shepherd, D, Ritter, G, Schmidt, R.R, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains.
Nat. Immunol., 3, 2002
|
|
1GZQ
 
 | | CD1b in complex with Phophatidylinositol | | Descriptor: | 2-[(HYDROXY{[(2R,3R,5S,6R)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL]OXY}PHOSPHORYL)OXY]-1-[(PALMITOYLOXY)METHYL]ETHYL HEPTADECANOATE, B2-MICROGLOBULIN, DOCOSANE, ... | | Authors: | Gadola, S.D, Zaccai, N.R, Harlos, K, Shepherd, D, Ritter, G, Schmidt, R.R, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-07-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of Human Cd1B with Bound Ligands at 2.3 A, a Maze for Alkyl Chains
Nat.Immunol., 3, 2002
|
|
1AGB
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGRKKYKL-3R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGRKKYKL - 3R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
3ZMN
 
 | | VP17, a capsid protein of bacteriophage P23-77 | | Descriptor: | VP17 | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
1BOY
 
 | |
6FKM
 
 | |
6F8P
 
 | |
6FKK
 
 | | Drosophila Semaphorin 1b, extracellular domains 1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MIP07328p, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rozbesky, D, Harlos, K, Jones, E.Y. | | Deposit date: | 2018-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
1CVW
 
 | | Crystal structure of active site-inhibited human coagulation factor VIIA (DES-GLA) | | Descriptor: | CALCIUM ION, COAGULATION FACTOR VIIA (HEAVY CHAIN) (DES-GLA), COAGULATION FACTOR VIIA (LIGHT CHAIN) (DES-GLA), ... | | Authors: | Kemball-Cook, G, Johnson, D.J.D, Tuddenham, E.G.D, Harlos, K. | | Deposit date: | 1999-08-24 | | Release date: | 1999-08-31 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of active site-inhibited human coagulation factor VIIa (des-Gla).
J.Struct.Biol., 127, 1999
|
|
4AE2
 
 | | Crystal structure of Human fibrillar procollagen type III C- propeptide trimer | | Descriptor: | CALCIUM ION, COLLAGEN ALPHA-1(III) CHAIN, NITRATE ION | | Authors: | Bourhis, J.M, Mariano, N, Zhao, Y, Harlos, K, Jones, E.Y, Moali, C, Aghajari, N, Hulmes, D.J. | | Deposit date: | 2012-01-05 | | Release date: | 2012-09-12 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis of Fibrillar Collagen Trimerization and Related Genetic Disorders.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3ZN4
 
 | | VP16, a capsid protein of bacteriophage P23-77 (VP16-type-2) | | Descriptor: | CHLORIDE ION, CITRIC ACID, VP16 | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
3ZN6
 
 | | VP16-VP17 complex, a complex of the two major capsid proteins of bacteriophage P23-77 | | Descriptor: | CHLORIDE ION, SODIUM ION, VP16, ... | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
4AEJ
 
 | | Crystal structure of Human fibrillar procollagen type III C- propeptide trimer | | Descriptor: | CALCIUM ION, COLLAGEN ALPHA-1(III) CHAIN, GLYCEROL | | Authors: | Bourhis, J.M, Mariano, N, Zhao, Y, Harlos, K, Jones, E.Y, Moali, C, Aghajari, N, Hulmes, D.J.S. | | Deposit date: | 2012-01-11 | | Release date: | 2012-09-12 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis of Fibrillar Collagen Trimerization and Related Genetic Disorders.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1DRY
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II), 2-OXOGLUTARATE AND N-ALPHA-L-ACETYL ARGININE | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, FE (II) ION, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1A1O
 
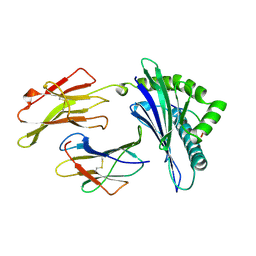 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE LS6 (KPIVQYDNF) FROM THE MALARIA PARASITE P. FALCIPARUM | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
5ZH3
 
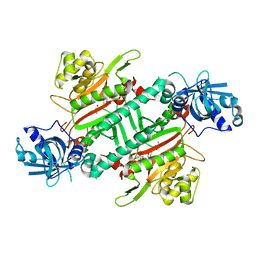 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-6 | | Descriptor: | (3S)-6,8-dihydroxy-3-{[(2R,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, LYSINE, Lysine-tRNA ligase | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
1A1M
 
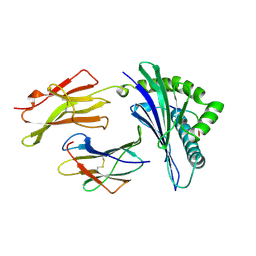 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE TPYDINQML FROM GAG PROTEIN OF HIV2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
5ZKF
 
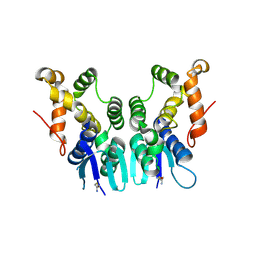 | | Crystal Structure of N-terminal Domain of Plasmodium vivax p43 in space group P21 | | Descriptor: | Aminoacyl tRNA Synthetase Complex-Interacting Multifunctional Protein p43 | | Authors: | Gupta, S, Sharma, M, Harlos, K, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-24 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the two domains that constitute the Plasmodium vivax p43 protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5ZKE
 
 | | Crystal Structure of N-terminal Domain of Plasmodium vivax p43 in space group P212121 | | Descriptor: | Aminoacyl tRNA Synthetase Complex-Interacting Multifunctional Protein p43 | | Authors: | Gupta, S, Sharma, M, Harlos, K, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Crystal structures of the two domains that constitute the Plasmodium vivax p43 protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5ZH5
 
 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-2 | | Descriptor: | (3S)-6,8-dihydroxy-3-{[(2R,6S)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, CHLORIDE ION, LYSINE, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
1DS1
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II) AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, FE (II) ION, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1DRT
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II), 2-OXOGLUTARATE AND PROCLAVAMINIC ACID | | Descriptor: | 2-OXOGLUTARIC ACID, 5-AMINO-3-HYDROXY-2-(2-OXO-AZETIDIN-1-YL)-PENTANOIC ACID, CLAVAMINATE SYNTHASE 1, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
