7BR3
 
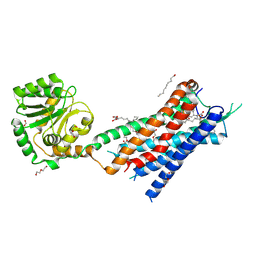 | | Crystal structure of the protein 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl dodecanoate, 4-[[(1R)-2-[5-(2-fluoranyl-3-methoxy-phenyl)-3-[[2-fluoranyl-6-(trifluoromethyl)phenyl]methyl]-4-methyl-2,6-bis(oxidanylidene)pyrimidin-1-yl]-1-phenyl-ethyl]amino]butanoic acid, ... | | Authors: | Cheng, L, Shao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode.
Nat Commun, 11, 2020
|
|
3ZIF
 
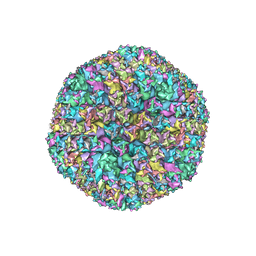 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | Descriptor: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | Authors: | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
3IZ3
 
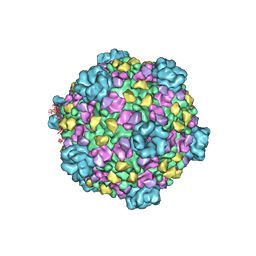 | | CryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Structural protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Cheng, L, Sun, J, Zhang, K, Mou, Z, Huang, X, Ji, G, Sun, F, Zhang, J, Zhu, P. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5Z4D
 
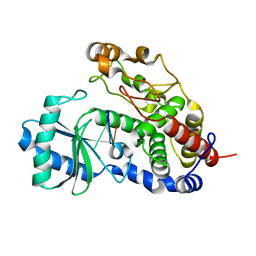 | | Structure of Tailor in complex with AGUU RNA | | Descriptor: | RNA (5'-R(*AP*GP*UP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4M
 
 | | Structure of TailorD343A with bound UTP and Mg | | Descriptor: | MAGNESIUM ION, Terminal uridylyltransferase Tailor, URIDINE 5'-TRIPHOSPHATE | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4C
 
 | | Crystal structure of Tailor | | Descriptor: | Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4J
 
 | | Structure of Tailor in complex with U4 RNA | | Descriptor: | RNA (5'-R(*UP*UP*UP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4A
 
 | | Structure of Tailor in complex with AGU RNA | | Descriptor: | RNA (5'-R(*AP*GP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.637 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
7ECA
 
 | |
4U6V
 
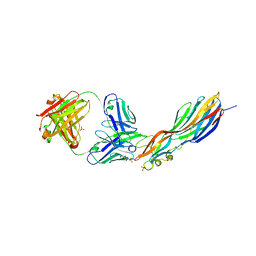 | | Mechanisms of Neutralization of a Human Anti-Alpha Toxin Antibody | | Descriptor: | Alpha-hemolysin, Fab, antigen binding fragment, ... | | Authors: | Oganesyan, V.Y, Peng, L, Damschroder, M.M, Cheng, L, Sadowska, A, Tkaczyk, C, Sellman, B, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2014-07-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Mechanisms of Neutralization of a Human Anti-alpha-toxin Antibody.
J.Biol.Chem., 289, 2014
|
|
4CIK
 
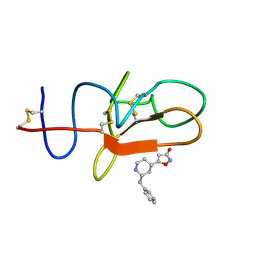 | | plasminogen kringle 1 in complex with inhibitor | | Descriptor: | 5-[(2R,4S)-2-(phenylmethyl)piperidin-4-yl]-1,2-oxazol-3-one, PLASMINOGEN | | Authors: | Xue, Y, Johansson, C, Cheng, L, Pettersen, D, Gustafsson, D. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of the Fibrinolysis Inhibitor Azd6564, Acting Via Interference of a Protein-Protein Interaction.
Acs Med.Chem.Lett., 5, 2014
|
|
4X9B
 
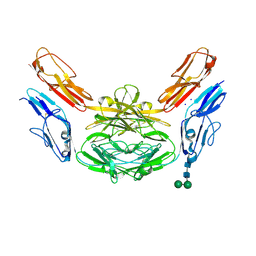 | | Crystal structure of Dscam1 isoform 4.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.44, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9G
 
 | | Crystal structure of Dscam1 isoform 6.44, N-terminal four Ig domains | | Descriptor: | Down Syndrome Cell Adhesion Molecule isoform 6.44, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X5L
 
 | | Crystal structure of Dscam1 Ig7 domain, isoform 9 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AM, SODIUM ION | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-05 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4XB7
 
 | | Crystal structure of Dscam1 isoform 4.4, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.4, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9I
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, isoform 9.44, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4XB8
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains (with zinc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4WVR
 
 | | Crystal structure of Dscam1 Ig7 domain, isoform 5 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AK | | Authors: | Chen, Q, Yu, Y, Li, S, Cheng, L. | | Deposit date: | 2014-11-07 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9F
 
 | | Crystal structure of Dscam1 isoform 6.9, N-terminal four Ig domains | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Down Syndrome Cell Adhesion Molecule isoform 6.9, GLYCEROL, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X83
 
 | | Crystal structure of Dscam1 isoform 7.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
3ERD
 
 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH DIETHYLSTILBESTROL AND A GLUCOCORTICOID RECEPTOR INTERACTING PROTEIN 1 NR BOX II PEPTIDE | | Descriptor: | ACETIC ACID, CHLORIDE ION, DIETHYLSTILBESTROL, ... | | Authors: | Shiau, A.K, Barstad, D, Loria, P.M, Cheng, L, Kushner, P.J, Agard, D.A, Greene, G.L. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.
Cell(Cambridge,Mass.), 95, 1998
|
|
3ERT
 
 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH 4-HYDROXYTAMOXIFEN | | Descriptor: | 4-HYDROXYTAMOXIFEN, PROTEIN (ESTROGEN RECEPTOR ALPHA) | | Authors: | Shiau, A.K, Barstad, D, Loria, P.M, Cheng, L, Kushner, P.J, Agard, D.A, Greene, G.L. | | Deposit date: | 1999-03-30 | | Release date: | 1999-04-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.
Cell(Cambridge,Mass.), 95, 1998
|
|
4RN5
 
 | | B1 domain of human Neuropilin-1 with acetate ion in a ligand-binding site | | Descriptor: | ACETATE ION, GLYCEROL, Neuropilin-1, ... | | Authors: | Allerston, C.K, Yelland, T.S, Jarvis, A, Jenkins, K, Winfield, N, Cheng, L, Jia, H, Zachary, I, Selwood, D.L, Djordjevic, S. | | Deposit date: | 2014-10-23 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Conserved water molecules in a ligand-binding site of neuropilin-1
To be Published
|
|
5H0S
 
 | | EM Structure of VP1A and VP1B | | Descriptor: | VP1 | | Authors: | Li, X, Zhou, N, Xu, B, Chen, W, Zhu, B, Wang, X, Wang, J, Liu, H, Cheng, L. | | Deposit date: | 2016-10-06 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
5H0R
 
 | | RNA dependent RNA polymerase ,vp4,dsRNA | | Descriptor: | RNA (42-MER), RNA-dependent RNA polymerase, VP4 protein | | Authors: | Li, X, Zhou, N, Chen, W, Zhu, B, Wang, X, Xu, B, Wang, J, Liu, H, Cheng, L. | | Deposit date: | 2016-10-06 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
