1MN9
 
 | | NDP kinase mutant (H122G) complex with RTP | | Descriptor: | MAGNESIUM ION, NDP kinase, RIBAVIRIN TRIPHOSPHATE | | Authors: | Gallois-montbrun, S, Chen, Y, Dutartre, H, Morera, S, Guerreiro, C, Mulard, L, Schneider, B, Janin, J, Canard, B, Veron, M, Deville-bonne, D. | | Deposit date: | 2002-09-05 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Analysis of the Activation of Ribavirin Analogs by NDP Kinase: Comparison with Other Ribavirin Targets
MOL.PHARMACOL., 63, 2003
|
|
4UD0
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UCY
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, SULFATE ION, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-18 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
1F3F
 
 | | STRUCTURE OF THE H122G NUCLEOSIDE DIPHOSPHATE KINASE / D4T-TRIPHOSPHATE.MG COMPLEX | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-DIPHOSPHATE, 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Meyer, P, Schneider, B, Sarfati, S, Deville-Bonne, D, Guerreiro, C, Boretto, J, Janin, J, Veron, M, Canard, B. | | Deposit date: | 2000-06-02 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase.
EMBO J., 19, 2000
|
|
4UCK
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UCZ
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | GUANOSINE, GUANOSINE-5'-TRIPHOSPHATE, RNA-DIRECTED RNA POLYMERASE L, ... | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UCL
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, SULFATE ION, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
1F6T
 
 | | STRUCTURE OF THE NUCLEOSIDE DIPHOSPHATE KINASE/ALPHA-BORANO(RP)-TDP.MG COMPLEX | | Descriptor: | 2*-DEOXY-THYMIDINE-5*-ALPHA BORANO DIPHOSPHATE (ISOMER RP), MAGNESIUM ION, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE) | | Authors: | Guerreiro, C, Boretto, J, Janin, J, Veron, M, Canard, B, Schneider, B, Sarfati, S, Deville-Bonne, D. | | Deposit date: | 2000-06-23 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase.
EMBO J., 19, 2000
|
|
3CDU
 
 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
3CDW
 
 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with protein primer VPg and a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
4UCJ
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, SULFATE ION, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
2J7U
 
 | | Dengue virus NS5 RNA dependent RNA polymerase domain | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA DEPENDENT RNA POLYMERASE, ... | | Authors: | Yap, T.L, Xu, T, Chen, Y.L, Malet, H, Egloff, M.P, Canard, B, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Dengue Virus RNA- Dependent RNA Polymerase Catalytic Domain at 1.85 Angstrom Resolution.
J.Virol., 81, 2007
|
|
2J7W
 
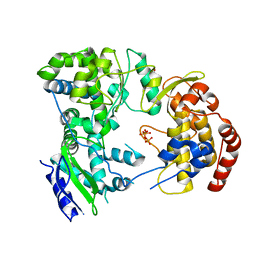 | | Dengue virus NS5 RNA dependent RNA polymerase domain complexed with 3' dGTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-TRIPHOSPHATE, POLYPROTEIN, ... | | Authors: | Yap, T.L, Xu, T, Chen, Y.L, Malet, H, Egloff, M.P, Canard, B, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Dengue Virus RNA- Dependent RNA Polymerase Catalytic Domain at 1.85 Angstrom Resolution.
J.Virol., 81, 2007
|
|
4UCI
 
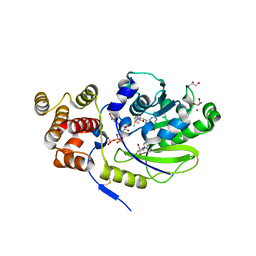 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | ADENOSINE, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UD1
 
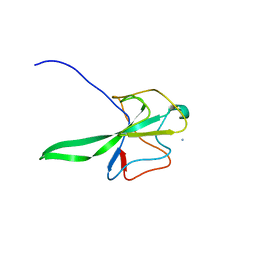 | | Structure of the N Terminal domain of the MERS CoV nucleocapsid | | Descriptor: | AMMONIUM ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Papageorgiou, N, Lichiere, J, Ferron, F, Canard, B, Coutard, B. | | Deposit date: | 2014-12-05 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Characterization of the N-Terminal Part of the Mers-Cov Nucleocapsid by X-Ray Diffraction and Small-Angle X-Ray Scattering
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5LS4
 
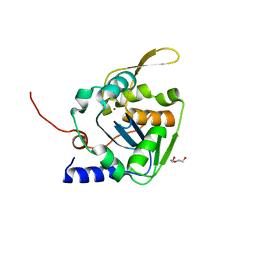 | | Mopeia virus exonuclease domain complexed with Calcium | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Nucleoprotein, ... | | Authors: | Yekwa, E.L, Canard, B, Ferron, F. | | Deposit date: | 2016-08-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.469 Å) | | Cite: | Activity inhibition and crystal polymorphism induced by active-site metal swapping.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2H85
 
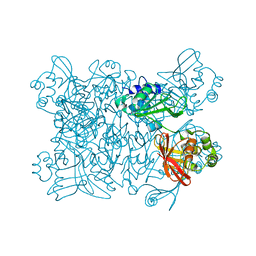 | | Crystal Structure of Nsp 15 from SARS | | Descriptor: | Putative orf1ab polyprotein | | Authors: | Ricagno, S, Egloff, M.P, Ulferts, R, Coutard, B, Nurizzo, D, Campanacci, V, Cambillau, C, Ziebuhr, J, Canard, B. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5LTF
 
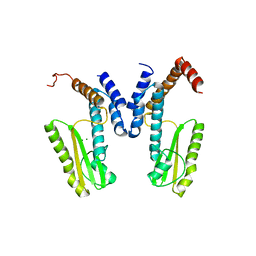 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease complexed with catalytic ions | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Ferron, F. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structures ofLymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
IUCrJ, 5, 2018
|
|
7P27
 
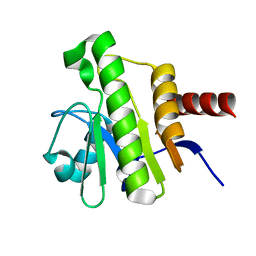 | | NMR solution structure of Chikungunya virus macro domain | | Descriptor: | Polyprotein P1234 | | Authors: | Lykouras, M.V, Tsika, A.C, Papageorgiou, N, Canard, B, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2021-07-04 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
2CKW
 
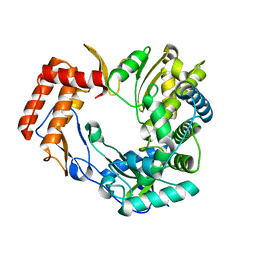 | | The 2.3 A resolution structure of the Sapporo virus RNA dependant RNA polymerase. | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Fullerton, S.W.B, Tucker, P.A, Rohayem, J, Coutard, B, Gebhardt, J, Gorbalenya, A, Canard, B. | | Deposit date: | 2006-04-24 | | Release date: | 2006-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Sapovirus RNA-Dependent RNA Polymerase.
J.Virol., 81, 2007
|
|
3OV9
 
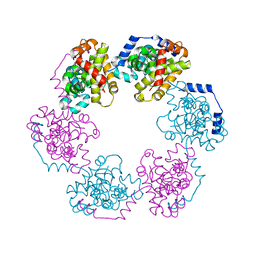 | | Structure of the Nucleoprotein from Rift Valley Fever Virus | | Descriptor: | NITRITE ION, Nucleoprotein, SODIUM ION | | Authors: | Ferron, F, Danek, E.I, Li, Z, Luo, D, Wong, Y.H, Coutard, B, Lantez, V, Charrel, R, Canard, B, Walz, T, Lescar, J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes
Plos Pathog., 7, 2011
|
|
3OUO
 
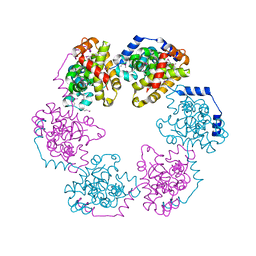 | | Structure of the Nucleoprotein from Rift Valley Fever Virus | | Descriptor: | NITRITE ION, Nucleoprotein | | Authors: | Ferron, F, Danek, E.I, Li, Z, Luo, D, Wong, Y.H, Coutard, B, Lantez, V, Charrel, R, Canard, B, Walz, T, Lescar, J. | | Deposit date: | 2010-09-15 | | Release date: | 2011-05-25 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes
Plos Pathog., 7, 2011
|
|
5LTS
 
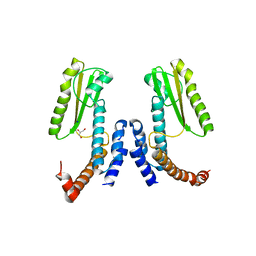 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease Mutant D118A | | Descriptor: | CHLORIDE ION, GLYCEROL, RNA-directed RNA polymerase L, ... | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Alvarez, K, Ferron, F. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures ofLymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
IUCrJ, 5, 2018
|
|
5NFY
 
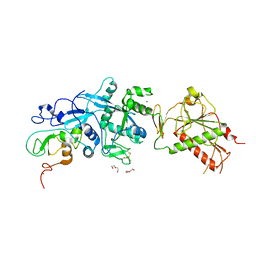 | | SARS-CoV nsp10/nsp14 dynamic complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, Polyprotein 1ab, ZINC ION | | Authors: | Ferron, F, Gluais, L, Vonrhein, C, Bricogne, G, Canard, B, Imbert, I. | | Deposit date: | 2017-03-16 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.382 Å) | | Cite: | Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5LTN
 
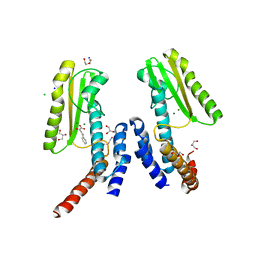 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease complexed with DPBA | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Alvarez, K, Ferron, F. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of Lymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
Iucrj, 5, 2018
|
|
