1DCH
 
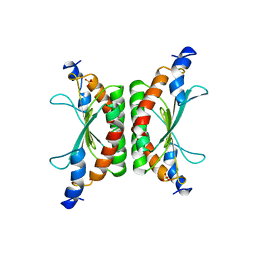 | | CRYSTAL STRUCTURE OF DCOH, A BIFUNCTIONAL, PROTEIN-BINDING TRANSCRIPTION COACTIVATOR | | Descriptor: | DCOH (DIMERIZATION COFACTOR OF HNF-1), SULFATE ION | | Authors: | Endrizzi, J.A, Cronk, J.D, Wang, W, Crabtree, G.R, Alber, T. | | Deposit date: | 1995-01-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of DCoH, a bifunctional, protein-binding transcriptional coactivator.
Science, 268, 1995
|
|
1DEB
 
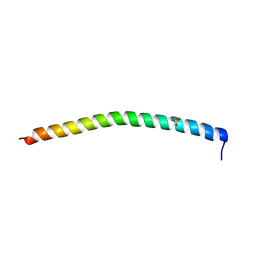 | |
1EKX
 
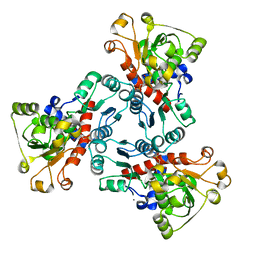 | | THE ISOLATED, UNREGULATED CATALYTIC TRIMER OF ASPARTATE TRANSCARBAMOYLASE COMPLEXED WITH BISUBSTRATE ANALOG PALA (N-(PHOSPHONACETYL)-L-ASPARTATE) | | Descriptor: | ASPARTATE TRANSCARBAMOYLASE, CALCIUM ION, N-(PHOSPHONACETYL)-L-ASPARTIC ACID | | Authors: | Endrizzi, J.A, Beernink, P.T, Alber, T, Schachman, H.K. | | Deposit date: | 2000-03-09 | | Release date: | 2000-05-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of bisubstrate analog promotes large structural changes in the unregulated catalytic trimer of aspartate transcarbamoylase: implications for allosteric regulation induced cell migration.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2O7A
 
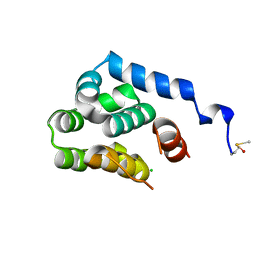 | | T4 lysozyme C-terminal fragment | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme | | Authors: | Echols, N, Kwon, E, Marqusee, S.M, Alber, T. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
2O60
 
 | |
2O5G
 
 | | Calmodulin-smooth muscle light chain kinase peptide complex | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Valentine, K.G, Ng, H.L, Schneeweis, J.K, Kranz, J.K, Frederick, K.K, Alber, T, Wand, A.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ultrahigh resolution crystal structure of calmodulin-smooth muscle light kinase peptide complex
To be Published
|
|
2O79
 
 | | T4 lysozyme with C-terminal extension | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme, ... | | Authors: | Llinas, M, Crowder, S.M, Echols, N, Alber, T, Marqusee, S. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
2O4W
 
 | | T4 lysozyme circular permutant | | Descriptor: | CHLORIDE ION, Lysozyme | | Authors: | Llinas, M, Crowder, S.M, Echols, N, Alber, T, Marqusee, S. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
2O6N
 
 | |
2OZ5
 
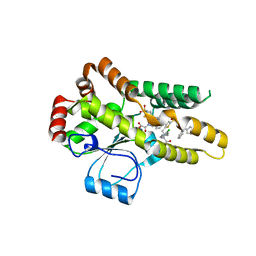 | | Crystal structure of Mycobacterium tuberculosis protein tyrosine phosphatase PtpB in complex with the specific inhibitor OMTS | | Descriptor: | Phosphotyrosine protein phosphatase ptpb, {(3-CHLOROBENZYL)[(5-{[(3,3-DIPHENYLPROPYL)AMINO]SULFONYL}-2-THIENYL)METHYL]AMINO}(OXO)ACETIC ACID | | Authors: | Grundner, C, Gee, C.L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Selective Inhibition of Mycobacterium tuberculosis Protein Tyrosine Phosphatase PtpB.
Structure, 15, 2007
|
|
3OUN
 
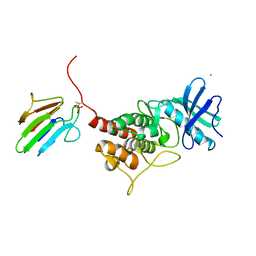 | |
3ORI
 
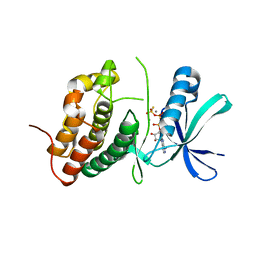 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 1) | | Descriptor: | MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Lombana, T.N, Echols, N, Good, M.C, Thomsen, N.D, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB.
Structure, 18, 2010
|
|
3OUK
 
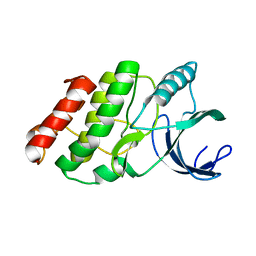 | |
3ORL
 
 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 3) | | Descriptor: | MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Echols, N, Lombana, T.N, Thomsen, N.D, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr kinase, PknB
Structure, 18, 2010
|
|
3ORP
 
 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 5) | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Good, M.C, Echols, N, Lombana, T.N, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr kinase, PknB
Structure, 18, 2010
|
|
3OTV
 
 | |
3ORO
 
 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 4) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Good, M.C, Echols, N, Lombana, T.N, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB.
Structure, 18, 2010
|
|
3OXH
 
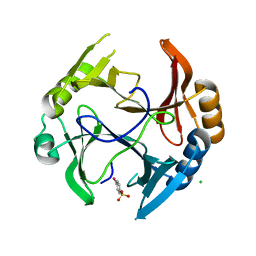 | | Mycobacterium tuberculosis kinase inhibitor homolog RV0577 | | Descriptor: | CHLORIDE ION, PARA-MERCURY-BENZENESULFONIC ACID, RV0577 PROTEIN, ... | | Authors: | Echols, N, Flynn, E.M, Stephenson, S, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-21 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biophysical Characterization of the Mycobacterium tuberculosis Protein Rv0577, a Protein Associated with Neutral Red Staining of Virulent Tuberculosis Strains and Homologue of the Streptomyces coelicolor Protein KbpA.
Biochemistry, 2017
|
|
3ORK
 
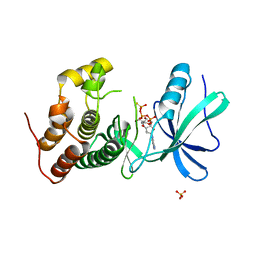 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Echols, N, Lombana, T.N, Thomsen, N.D, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr kinase, PknB
Structure, 18, 2010
|
|
3ORM
 
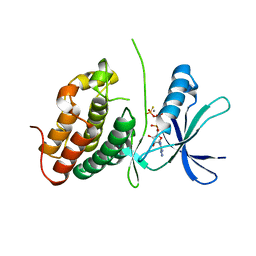 | | Mycobacterium tuberculosis PknB kinase domain D76A mutant | | Descriptor: | MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Echols, N, Lombana, T.N, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB.
Structure, 18, 2010
|
|
3ORT
 
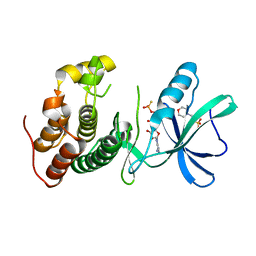 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 6) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Good, M.C, Echols, N, Lombana, T.N, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr kinase, PknB
Structure, 18, 2010
|
|
3OUV
 
 | |
3RMR
 
 | |
1RWL
 
 | | Extracellular domain of Mycobacterium tuberculosis PknD | | Descriptor: | CADMIUM ION, Serine/threonine-protein kinase pknD | | Authors: | Good, M.C, Greenstein, A.E, Young, T.A, Ng, H.L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sensor Domain of the Mycobacterium tuberculosis Receptor Ser/Thr Protein Kinase, PknD, forms a Highly Symmetric beta Propeller.
J.Mol.Biol., 339, 2004
|
|
1QSC
 
 | | CRYSTAL STRUCTURE OF THE TRAF DOMAIN OF TRAF2 IN A COMPLEX WITH A PEPTIDE FROM THE CD40 RECEPTOR | | Descriptor: | CD40 RECEPTOR, TNF RECEPTOR ASSOCIATED FACTOR 2 | | Authors: | McWhirter, S.M, Pullen, S.S, Holton, J.M, Crute, J.J, Kehry, M.R, Alber, T. | | Deposit date: | 1999-06-20 | | Release date: | 1999-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic analysis of CD40 recognition and signaling by human TRAF2.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
