4IJ7
 
 | | Crystal structure of Odorant Binding Protein 48 from Anopheles gambiae (AgamOBP48) with PEG | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Odorant binding protein-8, SODIUM ION | | Authors: | Zographos, S.E, Tsitsanou, K.E, Drakou, C.E. | | Deposit date: | 2012-12-21 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal and Solution Studies of the "Plus-C" Odorant-binding Protein 48 from Anopheles gambiae: CONTROL OF BINDING SPECIFICITY THROUGH THREE-DIMENSIONAL DOMAIN SWAPPING.
J.Biol.Chem., 288, 2013
|
|
3IIJ
 
 | |
3IIM
 
 | | The structure of hCINAP-dADP complex at 2.0 angstroms resolution | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zographos, S.E, Drakou, C.E, Leonidas, D.D. | | Deposit date: | 2009-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | hCINAP is an atypical mammalian nuclear adenylate kinase with an ATPase motif: Structural and functional studies.
Proteins, 80, 2012
|
|
3IIK
 
 | |
3IIL
 
 | | The structure of hCINAP-MgADP-Pi complex at 2.0 angstroms resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Coilin-interacting nuclear ATPase protein, LITHIUM ION, ... | | Authors: | Zographos, S.E, Drakou, C.E, Leonidas, D.D. | | Deposit date: | 2009-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | hCINAP is an atypical mammalian nuclear adenylate kinase with an ATPase motif: Structural and functional studies.
Proteins, 80, 2012
|
|
2AMV
 
 | | THE STRUCTURE OF GLYCOGEN PHOSPHORYLASE B WITH AN ALKYL-DIHYDROPYRIDINE-DICARBOXYLIC ACID | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Zographos, S.E, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of glycogen phosphorylase b with an alkyldihydropyridine-dicarboxylic acid compound, a novel and potent inhibitor.
Structure, 5, 1997
|
|
2QN1
 
 | | Glycogen Phosphorylase b in complex with asiatic acid | | Descriptor: | Glycogen phosphorylase, muscle form, asiatic acid | | Authors: | Zographos, S.E, Leonidas, D.D, Alexacou, K.-M, Hayes, J, Oikonomakos, N.G. | | Deposit date: | 2007-07-17 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Naturally occurring pentacyclic triterpenes as inhibitors of glycogen phosphorylase: synthesis, structure-activity relationships, and X-ray crystallographic studies
J.Med.Chem., 51, 2008
|
|
2QN2
 
 | | Glycogen Phosphorylase b in complex with Maslinic Acid | | Descriptor: | Glycogen phosphorylase, muscle form, maslinic acid | | Authors: | Zographos, S.E, Leonidas, D.D, Alexacou, K.-M, Hayes, J, Oikonomakos, N.G. | | Deposit date: | 2007-07-17 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Naturally occurring pentacyclic triterpenes as inhibitors of glycogen phosphorylase: synthesis, structure-activity relationships, and X-ray crystallographic studies
J.Med.Chem., 51, 2008
|
|
5EL2
 
 | |
8C6G
 
 | |
8C68
 
 | |
8C6E
 
 | |
3D6Q
 
 | | The RNase A- 5'-Deoxy-5'-N-piperidinouridine complex | | Descriptor: | 1-(5-deoxy-5-piperidin-1-yl-alpha-L-arabinofuranosyl)pyrimidine-2,4(1H,3H)-dione, CITRATE ANION, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-05-20 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Morpholino, piperidino, and pyrrolidino derivatives of pyrimidine nucleosides as inhibitors of ribonuclease A: synthesis, biochemical, and crystallographic evaluation.
J.Med.Chem., 52, 2009
|
|
3DJO
 
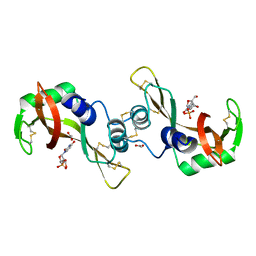 | | Bovine Seminal Ribonuclease uridine 2' phosphate complex | | Descriptor: | ACETATE ION, PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Seminal ribonuclease | | Authors: | Dossi, K, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the ribonucleolytic active site of bovine seminal ribonuclease. The binding of pyrimidinyl phosphonucleotide inhibitors
Eur.J.Med.Chem., 44, 2009
|
|
8BXU
 
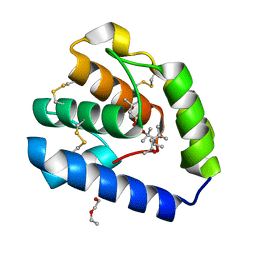 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with MPD (2-Methyl-2,4-pentanediol) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-ETHOXYETHANOL, Odorant binding protein, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-09 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
8BXV
 
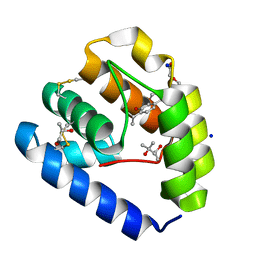 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with Thymol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-METHYL-2-(1-METHYLETHYL)PHENOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
8BY4
 
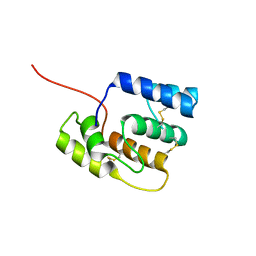 | |
8BXW
 
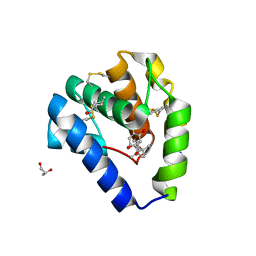 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with Carvacrol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-BUTANOL, 2-methyl-5-propan-2-yl-phenol, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
2GPA
 
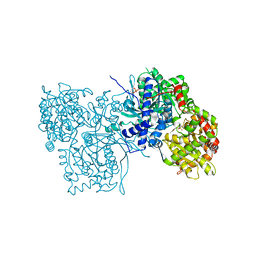 | | ALLOSTERIC INHIBITION OF GLYCOGEN PHOSPHORYLASE A BY A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oikonomakos, N.G, Tsitsanou, K.E, Zographos, S.E, Skamnaki, V.T. | | Deposit date: | 1999-02-18 | | Release date: | 1999-02-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of glycogen phosphorylase a by the potential antidiabetic drug 3-isopropyl 4-(2-chlorophenyl)-1,4-dihydro-1-ethyl-2-methyl-pyridine-3,5,6-tricarbo xylate.
Protein Sci., 8, 1999
|
|
2SKE
 
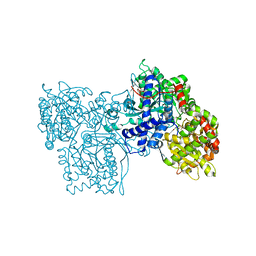 | | PYRIDOXAL PHOSPHORYLASE B IN COMPLEX WITH PHOSPHITE, GLUCOSE AND INOSINE-5'-MONOPHOSPHATE | | Descriptor: | INOSINIC ACID, PHOSPHITE ION, PYRIDOXAL PHOSPHORYLASE B, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Tsitsanou, K.E, Johnson, L.N, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Activator anion binding site in pyridoxal phosphorylase b: the binding of phosphite, phosphate, and fluorophosphate in the crystal.
Protein Sci., 5, 1996
|
|
2SKC
 
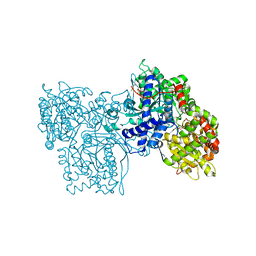 | | PYRIDOXAL PHOSPHORYLASE B IN COMPLEX WITH FLUOROPHOSPHATE, GLUCOSE AND INOSINE-5'-MONOPHOSPHATE | | Descriptor: | FLUORO-PHOSPHITE ION, INOSINIC ACID, PYRIDOXAL PHOSPHORYLASE B, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Tsitsanou, K.E, Johnson, L.N, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activator anion binding site in pyridoxal phosphorylase b: the binding of phosphite, phosphate, and fluorophosphate in the crystal.
Protein Sci., 5, 1996
|
|
2SKD
 
 | | PYRIDOXAL PHOSPHORYLASE B IN COMPLEX WITH PHOSPHATE, GLUCOSE AND INOSINE-5'-MONOPHOSPHATE | | Descriptor: | INOSINIC ACID, PHOSPHATE ION, PYRIDOXAL PHOSPHORYLASE B, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Tsitsanou, K.E, Johnson, L.N, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activator anion binding site in pyridoxal phosphorylase b: the binding of phosphite, phosphate, and fluorophosphate in the crystal.
Protein Sci., 5, 1996
|
|
1P4H
 
 | | Crystal structure of glycogen phosphorylase b in complex with C-(1-acetamido-alpha-D-glucopyranosyl) formamide | | Descriptor: | 1-DEOXY-1-ACETYLAMINO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Oikonomakos, N.G, Zographos, S.E, Kosmopoulou, M.N, Bischler, N, Leonidas, D.D, Nagy, V, Somsak, L, Gergely, P, Praly, J.-P. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crsytallographic Studies on alpha- and beta-D-glucopyranosyl Formamide Analogues, Inhibitors of Glycogen Phosphorylase
Biocatal.Biotransfor., 21, 2003
|
|
1P4J
 
 | | Crystal structure of glycogen phosphorylase b in complex with C-(1-hydroxy-beta-D-glucopyranosyl)formamide | | Descriptor: | (2R,3R,4S,5S,6R)-2,3,4,5-tetrahydroxy-6-(hydroxymethyl)oxane-2-carboxamide, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Oikonomakos, N.G, Zographos, S.E, Kosmopoulou, M.N, Bischler, N, Leonidas, D.D, Nagy, V, Somsak, L, Gergely, P, Praly, J.-P. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crsytallographic Studies on alpha- and beta-D-glucopyranosyl Formamide Analogues, Inhibitors of Glycogen Phosphorylase
Biocatal.Biotransfor., 21, 2003
|
|
1P4G
 
 | | Crystal structure of glycogen phosphorylase b in complex with C-(1-azido-alpha-D-glucopyranosyl)formamide | | Descriptor: | C-(1-AZIDO-ALPHA-D-GLUCOPYRANOSYL) FORMAMIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Oikonomakos, N.G, Zographos, S.E, Kosmopoulou, M.N, Bischler, N, Leonidas, D.D, Nagy, V, Somsak, L, Gergely, P, Praly, J.-P. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crsytallographic Studies on alpha- and beta-D-glucopyranosyl Formamide Analogues, Inhibitors of Glycogen Phosphorylase
Biocatal.Biotransfor., 21, 2003
|
|
