5O1Q
 
 | | LysF1 sh3b domain structure | | Descriptor: | sh3b domain | | Authors: | Benesik, M, Novacek, J, Janda, L, Dopitova, R, Pernisova, M, Melkova, K, Tisakova, L, Doskar, J, Zidek, L, Hejatko, J, Pantucek, R. | | Deposit date: | 2017-05-19 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity.
Virus Genes, 54, 2018
|
|
8BYR
 
 | |
8S8O
 
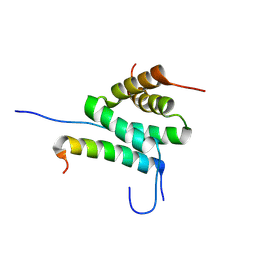 | | Solution Structure of cAMP-dependent Protein Kinase RII-alpha Subunit Dimerization and Docking Domain Complex with Microtubule Associated Protein 2c (84-111) | | Descriptor: | Isoform 3 of Microtubule-associated protein 2, cAMP-dependent protein kinase type II-alpha regulatory subunit | | Authors: | Bartosik, V, Lanikova, A, Janackova, Z, Padrta, P, Zidek, L. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-25 | | Method: | SOLUTION NMR | | Cite: | Structural basis of binding the unique N-terminal domain of microtubule-associated protein 2c to proteins regulating kinases of signaling pathways.
J.Biol.Chem., 300, 2024
|
|
6FVC
 
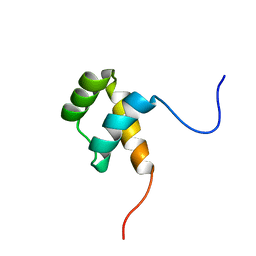 | | Protein environment affects the water-tryptophan binding mode. Molecular dynamics simulations of Engrailed homeodomain mutants | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Trosanova, Z, Zachrdla, M, Jansen, S, Srb, P, Zidek, L, Kozelka, J. | | Deposit date: | 2018-03-02 | | Release date: | 2019-04-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein environment affects the water-tryptophan binding mode. MD, QM/MM, and NMR studies of engrailed homeodomain mutants.
Phys Chem Chem Phys, 20, 2018
|
|
1PQT
 
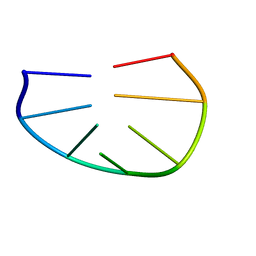 | |
2JNT
 
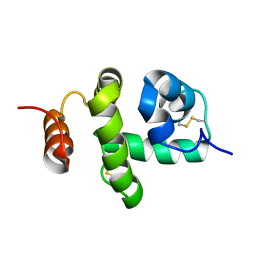 | | Structure of Bombyx mori Chemosensory Protein 1 in Solution | | Descriptor: | Chemosensory protein CSP1 | | Authors: | Jansen, S, Zidek, L, Chmelik, J, Novak, P, Padrta, P, Picimbon, J, Lofstedt, C, Sklenar, V. | | Deposit date: | 2007-02-02 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of Bombyx mori chemosensory protein 1 in solution
Arch.Insect Biochem.Physiol., 66, 2007
|
|
5LNM
 
 | | Crystal structure of D1050E mutant of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana | | Descriptor: | Histidine kinase CKI1 | | Authors: | Otrusinova, O, Demo, G, Kaderavek, P, Jansen, S, Jasenakova, Z, Pekarova, B, Janda, L, Wimmerova, M, Hejatko, J, Zidek, L. | | Deposit date: | 2016-08-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conformational dynamics are a key factor in signaling mediated by the receiver domain of a sensor histidine kinase from Arabidopsis thaliana.
J. Biol. Chem., 292, 2017
|
|
5LNN
 
 | | Crystal structure of D1050A mutant of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana | | Descriptor: | Histidine kinase CKI1 | | Authors: | Otrusinova, O, Demo, G, Kaderavek, P, Jansen, S, Jasenakova, Z, Pekarova, B, Janda, L, Wimmerova, M, Hejatko, J, Zidek, L. | | Deposit date: | 2016-08-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational dynamics are a key factor in signaling mediated by the receiver domain of a sensor histidine kinase from Arabidopsis thaliana.
J. Biol. Chem., 292, 2017
|
|
5MWW
 
 | | Sigma1.1 domain of sigmaA from Bacillus subtilis | | Descriptor: | RNA polymerase sigma factor SigA | | Authors: | Zachrdla, M, Padrta, P, Rabatinova, A, Sanderova, H, Barvik, I, Krasny, L, Zidek, L. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of domain 1.1 of the sigma (A) factor from Bacillus subtilis is preformed for binding to the RNA polymerase core.
J. Biol. Chem., 292, 2017
|
|
5N2N
 
 | | Crystal structure of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana complexed with Mg2+ and BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Histidine kinase CKI1, MAGNESIUM ION | | Authors: | Otrusinova, O, Demo, G, Padrta, P, Jasenakova, Z, Pekarova, B, Gelova, Z, Szmitkowska, A, Kaderavek, P, Jansen, S, Zachrdla, M, Klumpler, T, Marek, J, Hritz, J, Janda, L, Iwai, H, Wimmerova, M, Hejatko, J, Zidek, L. | | Deposit date: | 2017-02-08 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational dynamics are a key factor in signaling mediated by the receiver domain of a sensor histidine kinase from Arabidopsis thaliana.
J. Biol. Chem., 292, 2017
|
|
6SJX
 
 | | Precursor structure of the self-processing module of iron-regulated FrpC of N. Meningitidis with calcium ions | | Descriptor: | CALCIUM ION, Iron-regulated protein FrpC | | Authors: | Kuban, V, Macek, P, Hritz, J, Nechvatalova, K, Nedbalcova, K, Faldyna, M, Zidek, L, Bumba, L. | | Deposit date: | 2019-08-14 | | Release date: | 2020-02-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Ca 2+ -Dependent Self-Processing Activity of Repeat-in-Toxin Proteins.
Mbio, 11, 2020
|
|
6SJW
 
 | | Structure of the self-processing module of iron-regulated FrpC of N. Meningitidis with calcium ions | | Descriptor: | CALCIUM ION, Iron-regulated protein FrpC | | Authors: | Kuban, V, Macek, P, Hritz, J, Nechvatalova, K, Nedbalcova, K, Faldyna, M, Zidek, L, Bumba, L. | | Deposit date: | 2019-08-14 | | Release date: | 2020-02-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Ca 2+ -Dependent Self-Processing Activity of Repeat-in-Toxin Proteins.
Mbio, 11, 2020
|
|
3MM4
 
 | | Crystal structure of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana | | Descriptor: | Histidine kinase homolog | | Authors: | Marek, J, Klumpler, T, Pekarova, B, Triskova, O, Horak, J, Zidek, L, Dopitova, R, Hejatko, J, Janda, L. | | Deposit date: | 2010-04-19 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and binding specificity of the receiver domain of sensor histidine kinase CKI1 from Arabidopsis thaliana.
Plant J., 67, 2011
|
|
3MMN
 
 | | Crystal structure of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana complexed with Mg2+ | | Descriptor: | Histidine kinase homolog, MAGNESIUM ION | | Authors: | Marek, J, Klumpler, T, Pekarova, B, Triskova, O, Horak, J, Zidek, L, Dopitova, R, Hejatko, J, Janda, L. | | Deposit date: | 2010-04-20 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and binding specificity of the receiver domain of sensor histidine kinase CKI1 from Arabidopsis thaliana.
Plant J., 67, 2011
|
|
2KGF
 
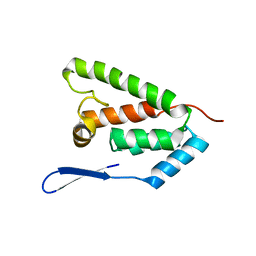 | | N-terminal domain of capsid protein from the Mason-Pfizer monkey virus | | Descriptor: | Capsid protein p27 | | Authors: | Macek, P, Chmelik, J, Zidek, L, Kaderavek, P, Padrta, P, Ruml, T, Pichova, I, Rumlova, M, Sklenar, V. | | Deposit date: | 2009-03-10 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of capsid protein from the mason-pfizer monkey virus
J.Mol.Biol., 392, 2009
|
|
4NC7
 
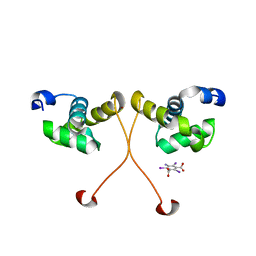 | | N-terminal domain of delta-subunit of RNA polymerase complexed with I3C and nickel ions | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, DNA-directed RNA polymerase subunit delta, NICKEL (II) ION | | Authors: | Demo, G, Papouskova, V, Komarek, J, Sanderova, H, Rabatinova, A, Krasny, L, Zidek, L, Sklenar, V, Wimmerova, M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray vs. NMR structure of N-terminal domain of delta-subunit of RNA polymerase.
J.Struct.Biol., 187, 2014
|
|
4NC8
 
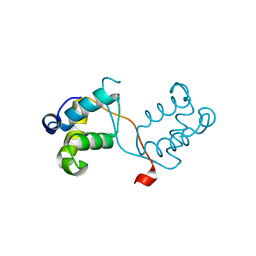 | | N-terminal domain of delta-subunit of RNA polymerase complexed with nickel ions | | Descriptor: | DNA-directed RNA polymerase subunit delta, NICKEL (II) ION | | Authors: | Demo, G, Papouskova, V, Komarek, J, Sanderova, H, Rabatinova, A, Krasny, L, Zidek, L, Sklenar, V, Wimmerova, M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | X-ray vs. NMR structure of N-terminal domain of delta-subunit of RNA polymerase.
J.Struct.Biol., 187, 2014
|
|
1I04
 
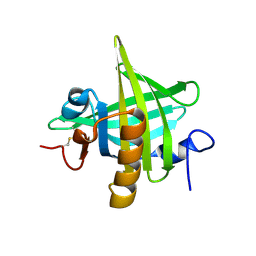 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN-I FROM MOUSE LIVER | | Descriptor: | MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of pheromone binding to mouse major urinary protein (MUP-I)
Protein Sci., 10, 2001
|
|
1I06
 
 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN (MUP-I) COMPLEXED WITH SEC-BUTYL-THIAZOLINE | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Pheromone Binding to Mouse Major Urinary Protein (MUP-I)
Protein Sci., 10, 2001
|
|
1I05
 
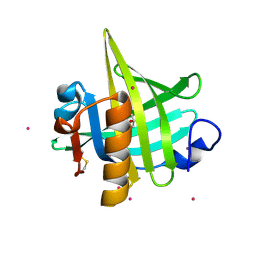 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN (MUP-I) COMPLEXED WITH HYDROXY-METHYL-HEPTANONE | | Descriptor: | 6-HYDROXY-6-METHYL-HEPTAN-3-ONE, CADMIUM ION, MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Pheromone Binding to Mouse Major Urinary Protein (MUP-I)
Protein Sci., 10, 2001
|
|
1KR8
 
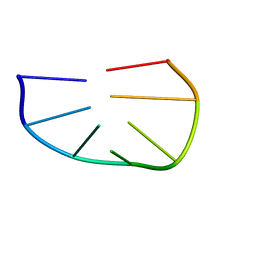 | |
2KRC
 
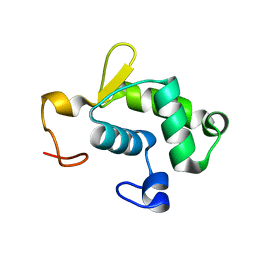 | | Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit delta | | Authors: | Motackova, V, Sanderova, H, Zidek, L, Novacek, J, Padrta, P, Svenkova, A, Jonak, J, Krasny, L, Sklenar, V. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase and its classification based on structural homologs
Proteins, 78, 2010
|
|
2M4K
 
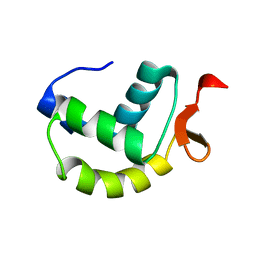 | | Solution structure of the delta subunit of RNA polymerase from Bacillus subtilis | | Descriptor: | DNA-directed RNA polymerase subunit delta | | Authors: | Papouskova, V, Novacek, J, Kaderavek, P, Zidek, L, Rabatinova, A, Sanderova, H, Krasny, L, Sklenar, V. | | Deposit date: | 2013-02-07 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Study of the Partially Disordered Full-Length delta Subunit of RNA Polymerase from Bacillus subtilis.
Chembiochem, 14, 2013
|
|
2LUF
 
 | |
