6J9M
 
 | | NmeBH+AcrIIC2 | | Descriptor: | AcrIIC2, CRISPR-associated endonuclease Cas9 | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6J9K
 
 | | Apo-AcrIIC2 | | Descriptor: | AcrIIC2, MAGNESIUM ION | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6J9L
 
 | | FnoBH+AcrIIC2 | | Descriptor: | AcrIIC2, HNH endonuclease family protein | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6J9N
 
 | | NmeHNH+AcrIIC3 | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9 | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
4NJL
 
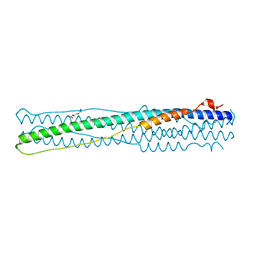 | | Crystal structure of middle east respiratory syndrome coronavirus S2 protein fusion core | | Descriptor: | S protein, TRIETHYLENE GLYCOL | | Authors: | Zhu, Y, Lu, L, Qin, L, Ye, S, Jiang, S, Zhang, R. | | Deposit date: | 2013-11-10 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor.
Nat Commun, 5, 2014
|
|
8I7O
 
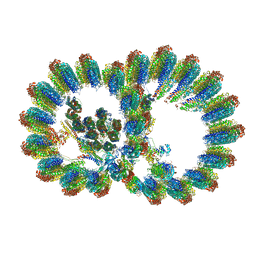 | | In situ structure of axonemal doublet microtubules in mouse sperm with 16-nm repeat | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 20, Cilia- and flagella-associated protein 276, ... | | Authors: | Zhu, Y, Yin, G.L, Tai, L.H, Sun, F. | | Deposit date: | 2023-02-01 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | In-cell structural insight into the stability of sperm microtubule doublet.
Cell Discov, 9, 2023
|
|
8I7R
 
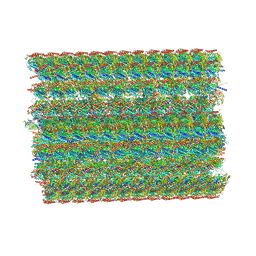 | | In situ structure of axonemal doublet microtubules in mouse sperm with 48-nm repeat | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Zhu, Y, Yin, G.L, Tai, L.H, Sun, F. | | Deposit date: | 2023-02-02 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | In-cell structural insight into the stability of sperm microtubule doublet.
Cell Discov, 9, 2023
|
|
3TG7
 
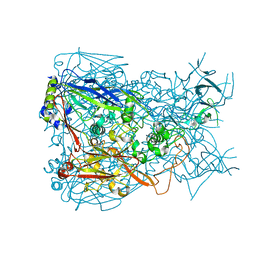 | | Crystal structure of Adenovirus serotype 5 hexon at 1.6A resolution | | Descriptor: | Hexon protein | | Authors: | Zhu, Y, Roszak, A.W, Isaacs, N.W, McVey, J.H, Nicklin, S.A, Baker, A.H. | | Deposit date: | 2011-08-17 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | crystal structure of Adenovirus serotype 5 hexon at 1.6A resolution
To be Published
|
|
2F5I
 
 | |
6JGY
 
 | | Crystal structure of LASV-GP2 in a post fusion conformation | | Descriptor: | Pre-glycoprotein polyprotein GP complex | | Authors: | Zhu, Y, Zhang, X, Chen, B, Ye, S, Zhang, R. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.389 Å) | | Cite: | Crystal Structure of Refolding Fusion Core of Lassa Virus GP2 and Design of Lassa Virus Fusion Inhibitors.
Front Microbiol, 10, 2019
|
|
5VFP
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFR
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFU
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFS
 
 | | Nucleotide-Driven Triple-State Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFT
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFQ
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFO
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-08 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
7FB3
 
 | |
7FB0
 
 | |
7FB1
 
 | |
7FB4
 
 | |
2WZS
 
 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 in complex with Mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhu, Y, Suits, M.D.L, Thompson, A, Chavan, S, Dinev, Z, Dumon, C, Smith, N, Moremen, K.W, Xiang, Y, Siriwardena, A, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-12-02 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
7WGZ
 
 | | SARS-CoV-2 spike glycoprotein trimer in open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
2G3T
 
 | |
7WVY
 
 | | Cryo-EM structure of the human formyl peptide receptor 2 in complex with Abeta42 and Gi2 | | Descriptor: | Amyloid-beta A4 protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, Lin, X, Zong, X, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-11 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of FPR2 in recognition of A beta 42 and neuroprotection by humanin.
Nat Commun, 13, 2022
|
|
