7WVU
 
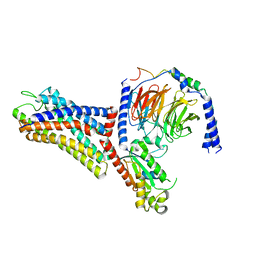 | | Cryo-EM structure of the human formyl peptide receptor 1 in complex with fMLF and Gi1 | | Descriptor: | FME-LEU-PHE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, Lin, X, Zong, X, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-11 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of FPR2 in recognition of A beta 42 and neuroprotection by humanin.
Nat Commun, 13, 2022
|
|
7WVV
 
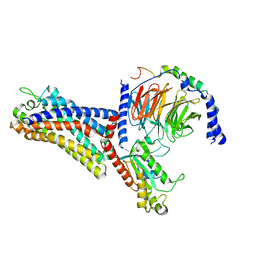 | | Cryo-EM structure of the human formyl peptide receptor 2 in complex with fMLFII and Gi2 | | Descriptor: | FME-LEU-PHE-ILE-ILE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, Lin, X, Zong, X, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-11 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of FPR2 in recognition of A beta 42 and neuroprotection by humanin.
Nat Commun, 13, 2022
|
|
7WGZ
 
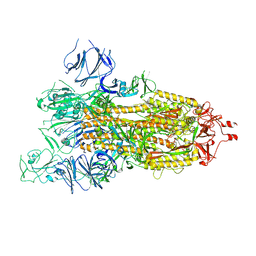 | | SARS-CoV-2 spike glycoprotein trimer in open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
6AKY
 
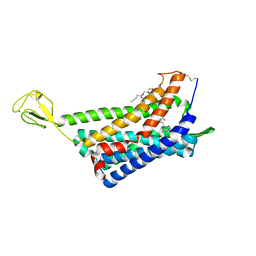 | | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 34 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]octan-8-yl}-1-(thiophen-3-yl)propyl]cyclohexane-1-carboxamide, C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, ... | | Authors: | Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists.
J. Med. Chem., 61, 2018
|
|
6AKX
 
 | | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 21 | | Descriptor: | C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]octan-8-yl}-1-(thiophen-2-yl)propyl]cyclopentanecarboxamide, NITRATE ION, ... | | Authors: | Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists.
J. Med. Chem., 61, 2018
|
|
7CWK
 
 | |
7F01
 
 | |
7FB1
 
 | |
7FB3
 
 | |
7FB0
 
 | |
7FB4
 
 | |
7Y37
 
 | |
8WR2
 
 | | Crystal Structure of Human Pyridoxal Kinase with bound Luteolin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Fan, J, Zhu, Y. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and characterization of natural product luteolin as an effective inhibitor of human pyridoxal kinase.
Bioorg.Chem., 143, 2024
|
|
8XEP
 
 | |
4UTF
 
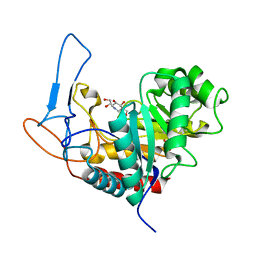 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-isofagomine and alpha- 1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, ... | | Authors: | Cuskin, F, Lowe, E.C, Temple, M.J, Zhu, Y, Pudlo, N.A, Cameron, E.A, Urs, K, Thompson, A.J, Cartmell, A, Rogowski, A, Tolbert, T, Piens, K, Bracke, D, Vervecken, W, Hakki, Z, Speciale, G, Munoz-Munoz, J.L, Pena, M.J, McLean, R, Suits, M.D, Boraston, A.B, Atherly, T, Ziemer, C.J, Williams, S.J, Davies, G.J, Abbott, D.W, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
1G9I
 
 | | CRYSTAL STRUCTURE OF BETA-TRYSIN COMPLEX IN CYCLOHEXANE | | Descriptor: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | Authors: | Zhu, G, Huang, Q, Zhu, Y, Li, Y, Chi, C, Tang, Y. | | Deposit date: | 2000-11-24 | | Release date: | 2000-12-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray study on an artificial mung bean inhibitor complex with bovine beta-trypsin in neat cyclohexane.
Biochim.Biophys.Acta, 1546, 2001
|
|
1X7I
 
 | | Crystal structure of the native copper homeostasis protein (cutCm) with calcium binding from Shigella flexneri 2a str. 301 | | Descriptor: | CALCIUM ION, Copper homeostasis protein cutC | | Authors: | Zhu, D.Y, Zhu, Y.Q, Huang, R.H, Xiang, Y, Wang, D.C. | | Deposit date: | 2004-08-14 | | Release date: | 2005-03-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the copper homeostasis protein (CutCm) from Shigella flexneri at 1.7 A resolution: The first structure of a new sequence family of TIM barrels
Proteins, 58, 2004
|
|
8JCJ
 
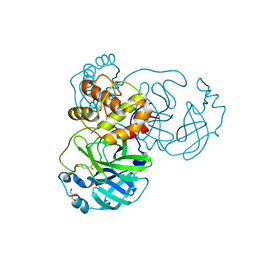 | |
8JCK
 
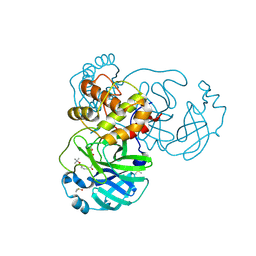 | |
8JCL
 
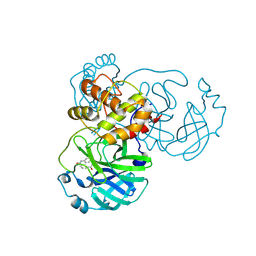 | |
8JCM
 
 | |
8JCN
 
 | |
8JCO
 
 | |
2WW0
 
 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | 1S-8AB-OCTAHYDRO-INDOLIZIDINE-1A,2A,8B-TRIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Suits, M.D.L, Thompson, A, Zhu, Y, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
2YG0
 
 | | CBM62 FROM CLOSTRIDIUM THERMOCELLUM XYL5A | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Montanier, C.Y, Correia, M.A.S, Flint, J.E, Zhu, Y, Basle, A, Mckee, L.S, Prates, J.A.M, Polizzi, S.J, Coutinho, P.M, Henrissat, B, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel, Noncatalytic Carbohydrate-Binding Module Displays Specificity for Galactose-Containing Polysaccharides Through Calcium-Mediated Oligomerization.
J.Biol.Chem., 286, 2011
|
|
