7ETU
 
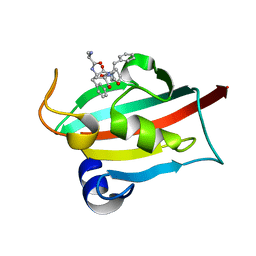 | | The FK1 domain of FKBP51 in complex with peptide-inhibitor hit SFPFT | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, peptide-inhibitor hit | | Authors: | Han, J.T, Zhu, Y.C, Pan, D.B, Xue, H.X, Wang, S, Liu, H.X, He, Y.X, Yao, X.J. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery of pentapeptide-inhibitor hits targeting FKBP51 by combining computational modeling and X-ray crystallography.
Comput Struct Biotechnol J, 19, 2021
|
|
7CRD
 
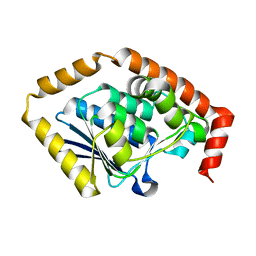 | | Structure of Pseudomonas aeruginosa OdaA | | Descriptor: | Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
7CBL
 
 | | Cryo-EM structure of the flagellar LP ring from Salmonella | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-12 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
5YEI
 
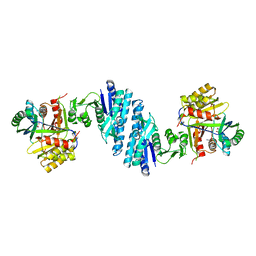 | | Mechanistic insight into the regulation of Pseudomonas aeruginosa aspartate kinase | | Descriptor: | Aspartokinase, GLYCEROL, LYSINE, ... | | Authors: | Li, C, Yang, M, Liu, L, Peng, C, Li, T, He, L, Song, Y, Zhu, Y, Bao, R. | | Deposit date: | 2017-09-17 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the allosteric regulation of Pseudomonas aeruginosa aspartate kinase.
Biochem.J., 475, 2018
|
|
7ENG
 
 | |
5XNB
 
 | |
7F1R
 
 | | Cryo-EM structure of the chemokine receptor CCR5 in complex with RANTES and Gi | | Descriptor: | C-C motif chemokine 5,C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7F1S
 
 | | Cryo-EM structure of the apo chemokine receptor CCR5 in complex with Gi | | Descriptor: | C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7F1Q
 
 | | Cryo-EM structure of the chemokine receptor CCR5 in complex with MIP-1a and Gi | | Descriptor: | C-C motif chemokine 3,C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7E9Q
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 out RBD, state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9O
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(3 up RBDs, state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENF
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7F8Y
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with devazepide | | Descriptor: | N-[(3S)-1-methyl-2-oxidanylidene-5-phenyl-3H-1,4-benzodiazepin-3-yl]-1H-indole-2-carboxamide, fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8U
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with lintitript | | Descriptor: | 2-[2-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]carbamoyl]indol-1-yl]ethanoic acid, Fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8W
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gq | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8V
 
 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gi | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8X
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with NN9056 | | Descriptor: | ASP-SMF-NLE-GLY-TRP-NLE-OEM-MEA-NH2 (NN9056), Cholecystokinin receptor type A,Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7FJD
 
 | | Cryo-EM structure of a membrane protein(WT) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJE
 
 | | Cryo-EM structure of a membrane protein(LL) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJF
 
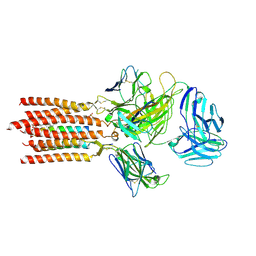 | | Cryo-EM structure of a membrane protein(CS) | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7F1T
 
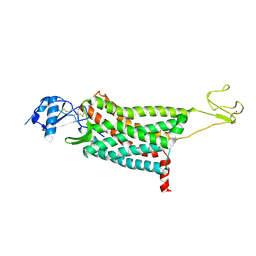 | | Crystal structure of the human chemokine receptor CCR5 in complex with MIP-1a | | Descriptor: | C-C motif chemokine 3,C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, ZINC ION | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7EBP
 
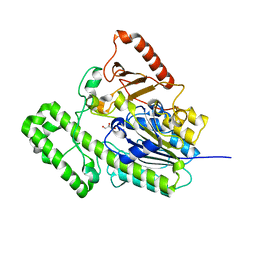 | | The structural analysis of A.Muciniphila sulfatase | | Descriptor: | CALCIUM ION, GLYCEROL, Sulfatase | | Authors: | Bao, R, Li, C.C, Tang, X.Y, Zhu, Y.B, Song, Y.J, Zhao, N.L, Huang, Q, Mou, X.Y, Luo, G.H, Liu, T.G. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.80000055 Å) | | Cite: | Structural analysis of the sulfatase AmAS from Akkermansia muciniphila.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EBQ
 
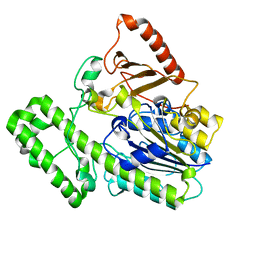 | | The structural analysis of A.Muciniphila sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Sulfatase | | Authors: | Bao, R, Li, C.C, Tang, X.Y, Zhu, Y.B, Song, Y.J, Zhao, N.L, Huang, Q, Mou, X.Y, Luo, G.H, Liu, T.G. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of the sulfatase AmAS from Akkermansia muciniphila.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EZT
 
 | | The structure and functional mechanism of nucleotide regulated acetylhexosaminidase Am2136 from Akkermansia muciniphila | | Descriptor: | Beta-N-acetylhexosaminidase, MAGNESIUM ION | | Authors: | Bao, R, Li, C.C, Tang, X.Y, Zhu, Y.B, Song, Y.J, Zhao, N.L, Huang, Q, Mou, X.Y, Luo, G.H, Liu, T.G. | | Deposit date: | 2021-06-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Nucleotide binding as an allosteric regulatory mechanism for Akkermansia muciniphila beta- N -acetylhexosaminidase Am2136.
Gut Microbes, 14, 2022
|
|
4HCN
 
 | | Crystal structure of Burkholderia pseudomallei effector protein CHBP in complex with ubiquitin | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Yao, Q, Cui, J, Zhu, Y, Shao, F. | | Deposit date: | 2012-09-30 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of ubiquitin and NEDD8 deamidation catalyzed by bacterial effectors that induce macrophage-specific apoptosis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
