7TP3
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K288.2 | | Descriptor: | CACODYLATE ION, K288.2 heavy chain, K288.2 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TP4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K398.22 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, K398.22 heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
5EQE
 
 | | Crystal structure of choline kinase alpha-1 bound by [4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]methanamine (compound 11) | | Descriptor: | Choline kinase alpha, [4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]methanamine | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
5EQP
 
 | | Crystal structure of choline kinase alpha-1 bound by 6-[(4-methyl-1,4-diazepan-1-yl)methyl]quinoline (compound 37) | | Descriptor: | 6-[(4-methyl-1,4-diazepan-1-yl)methyl]quinoline, Choline kinase alpha | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V2L
 
 | | Crystal structure of IRAK4 kinase domain with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, N-{2-[4-(hydroxymethyl)phenyl]-6-(2-hydroxypropan-2-yl)-2H-indazol-5-yl}-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
5FNV
 
 | | a new complex structure of tubulin with an alpha-beta unsaturated lactone | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, Y, Naismith, J, Zhu, X. | | Deposit date: | 2015-11-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Pironetin Reacts Covalently with Cysteine-316 of Alpha-Tubulin to Destabilize Microtubule.
Nat.Commun., 7, 2016
|
|
2QOH
 
 | | Crystal Structure of Abl kinase bound with PPY-A | | Descriptor: | 5-[3-(2-METHOXYPHENYL)-1H-PYRROLO[2,3-B]PYRIDIN-5-YL]-N,N-DIMETHYLPYRIDINE-3-CARBOXAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Dalgarno, D, Zhu, X. | | Deposit date: | 2007-07-20 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the T315I Mutant of Abl Kinase
Chem.Biol.Drug Des., 70, 2007
|
|
2WEU
 
 | |
2V7X
 
 | | X-RAY CRYSTAL STRUCTURE OF 5'-FLUORODEOXYADENOSINE SYNTHASE S158A mutant FROM STREPTOMYCES CATTLEYA COMPLEXED WITH the PRODUCTS, FDA and Met | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE, 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, METHIONINE | | Authors: | Robinson, D.A, Zhu, X, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2007-08-02 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mechanism of enzymatic fluorination in Streptomyces cattleya.
J. Am. Chem. Soc., 129, 2007
|
|
2WET
 
 | | Crystal structure of tryptophan 5-halogenase (PyrH) complex with FAD (tryptophan) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | De Laurentis, W, Zhu, X, Naismith, J.H. | | Deposit date: | 2009-04-01 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights in the Regioselectivity in the Enzymatic Chlorination of Tryptophan.
J.Mol.Biol., 391, 2009
|
|
4QC2
 
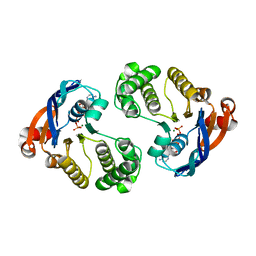 | | Crystal structure of lipopolysaccharide transport protein LptB in complex with ATP and Magnesium ions | | Descriptor: | ABC transporter related protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, Z, Xiang, Q, Zhu, X, Dong, H, He, C, Wang, H, Zhang, Y, Wang, W, Dong, C. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and functional studies of conserved nucleotide-binding protein LptB in lipopolysaccharide transport.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
7WZ7
 
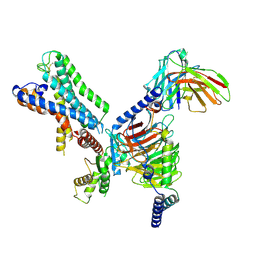 | | GPR110/G12 complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-17 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
7WXU
 
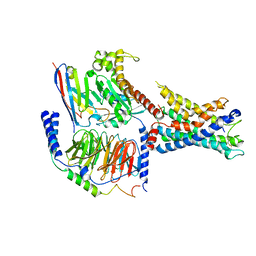 | | GPR110/Gq complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
7WY0
 
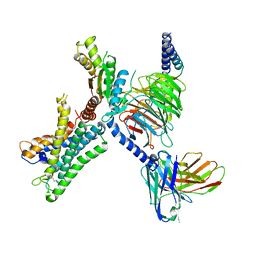 | | GPR110/G13 complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Engineered G alpha 13 subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
7WXW
 
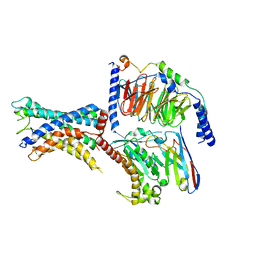 | | GPR110/Gs complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Engineered mini Galpha-s subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
7X2V
 
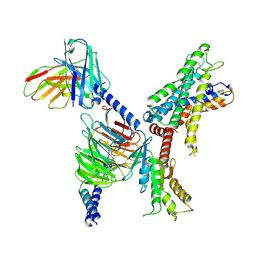 | | GPR110/Gi complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
5T0D
 
 | | Crystal structure of H6 hemagglutinin G225D mutant from Taiwan (2013) H6N1 influenza virus in complex with 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | Authors: | Wilson, I.A, Tzarum, N, Zhu, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | A single mutation in Taiwanese H6N1 influenza hemagglutinin switches binding to human-type receptors.
EMBO Mol Med, 9, 2017
|
|
5T08
 
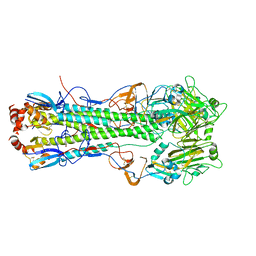 | |
5T0E
 
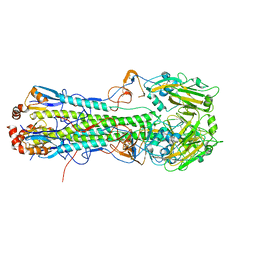 | | Crystal structure of H6 hemagglutinin G225D mutant from Taiwan (2013) H6N1 influenza virus in complex with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | Authors: | Wilson, I.A, Tzarum, N, Zhu, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | A single mutation in Taiwanese H6N1 influenza hemagglutinin switches binding to human-type receptors.
EMBO Mol Med, 9, 2017
|
|
7KN5
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobodies VHH E and U | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7KN7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobody VHH W and antibody Fab CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, CC12.3 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7JMO
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-04 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-04 heavy chain, COVA2-04 light chain, ... | | Authors: | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
7JMP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-39 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-39 heavy chain, COVA2-39 light chain, ... | | Authors: | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
