4I9H
 
 | | Crystal structure of rabbit LDHA in complex with AP28669 | | Descriptor: | 1-O-[3-(5-carboxypyridin-2-yl)-5-fluorophenyl]-6-O-[4-({[(5-carboxypyridin-2-yl)sulfanyl]acetyl}amino)-2-chloro-5-methoxyphenyl]-D-mannitol, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4I8X
 
 | | Crystal structure of rabbit LDHA in complex with AP27460 | | Descriptor: | 6-phenylpyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4I9N
 
 | | Crystal structure of rabbit LDHA in complex with AP28161 and AP28122 | | Descriptor: | 6-({2-[(5-chloro-4-{[(2S)-2,3-dihydroxypropyl]oxy}-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, 6-[3-(carboxymethoxy)-5-fluorophenyl]pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Kohlmann, A, Stephan, Z.G, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
8GB5
 
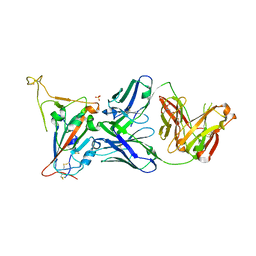 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 25F9 | | Descriptor: | 25F9 Heavy chain, 25F9 Light chain, BICINE, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-02-24 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Broadly neutralizing antibodies against sarbecoviruses generated by immunization of macaques with an AS03-adjuvanted COVID-19 vaccine.
Sci Transl Med, 15, 2023
|
|
5U36
 
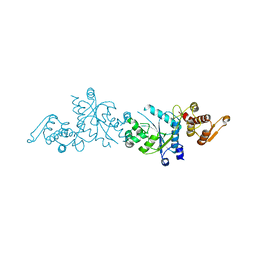 | | Crystal Structure Of A Mutant M. Jannashii Tyrosyl-tRNA Synthetase | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Luo, X, Fu, G, Zhu, X, Wilson, I.A, Wang, F. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria.
Nat. Chem. Biol., 13, 2017
|
|
5YDT
 
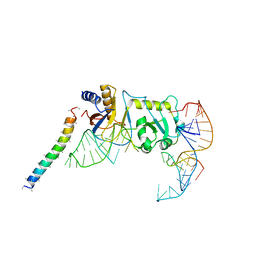 | | Remodeled Utp30 in 90S pre-ribosome (Mtr4-depleted, Enp1-TAP) | | Descriptor: | 5' ETS RNA, Ribosome biogenesis protein UTP30, Saccharomyces cerevisiae strain ALI 308 18S ribosomal RNA gene, ... | | Authors: | Ye, K, Zhu, X, Hu, J. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
5T0D
 
 | | Crystal structure of H6 hemagglutinin G225D mutant from Taiwan (2013) H6N1 influenza virus in complex with 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | Authors: | Wilson, I.A, Tzarum, N, Zhu, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | A single mutation in Taiwanese H6N1 influenza hemagglutinin switches binding to human-type receptors.
EMBO Mol Med, 9, 2017
|
|
5T08
 
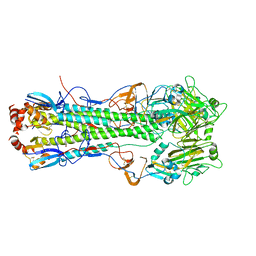 | |
5T0E
 
 | | Crystal structure of H6 hemagglutinin G225D mutant from Taiwan (2013) H6N1 influenza virus in complex with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | Authors: | Wilson, I.A, Tzarum, N, Zhu, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | A single mutation in Taiwanese H6N1 influenza hemagglutinin switches binding to human-type receptors.
EMBO Mol Med, 9, 2017
|
|
8GF2
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies eCR3022.20 and CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Broadening a SARS-CoV-1-neutralizing antibody for potent SARS-CoV-2 neutralization through directed evolution.
Sci.Signal., 16, 2023
|
|
5XGL
 
 | | Co-crystal structure of Ac-AChBPP in complex with alpha-conotoxin LvIA | | Descriptor: | Alpha-conotoxin LvIA, Soluble acetylcholine receptor | | Authors: | Wang, X.Q, Xu, M.Y, Luo, S.L, Zhu, X.P. | | Deposit date: | 2017-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.439 Å) | | Cite: | The crystal structure of Ac-AChBP in complex with alpha-conotoxin LvIA reveals the mechanism of its selectivity towards different nAChR subtypes
Protein Cell, 8, 2017
|
|
8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V2L
 
 | | Crystal structure of IRAK4 kinase domain with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, N-{2-[4-(hydroxymethyl)phenyl]-6-(2-hydroxypropan-2-yl)-2H-indazol-5-yl}-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
4U3F
 
 | | Cytochrome bc1 complex from chicken with designed inhibitor bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CARDIOLIPIN, ... | | Authors: | Huang, L.-S, Zhu, X.-L, Yang, G.F, Berry, E.A. | | Deposit date: | 2014-07-21 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2312 Å) | | Cite: | Rational Design of Highly Potent and Slow-Binding Cytochrome bc1 Inhibitor as Fungicide by Computational Substitution Optimization
Sci Rep, 5, 2015
|
|
2V7X
 
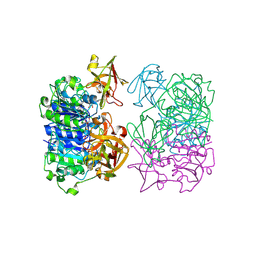 | | X-RAY CRYSTAL STRUCTURE OF 5'-FLUORODEOXYADENOSINE SYNTHASE S158A mutant FROM STREPTOMYCES CATTLEYA COMPLEXED WITH the PRODUCTS, FDA and Met | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE, 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, METHIONINE | | Authors: | Robinson, D.A, Zhu, X, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2007-08-02 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mechanism of enzymatic fluorination in Streptomyces cattleya.
J. Am. Chem. Soc., 129, 2007
|
|
5YDU
 
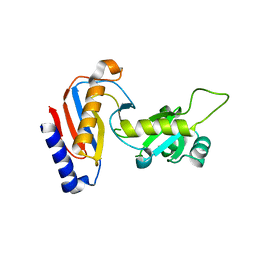 | | Crystal structure of Utp30 | | Descriptor: | PHOSPHATE ION, Ribosome biogenesis protein UTP30 | | Authors: | Hu, J, Zhu, X, Ye, K. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
5YJD
 
 | |
5YJC
 
 | |
6AGF
 
 | | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, X.J, li, Z.Q, Zhou, Q, Shen, H.Z, Wu, K, Huang, X.S, Chen, J.F, Zhang, J.R, Zhu, X.C, Lei, J.L, Xiong, W, Gong, H.P, Xiao, B.L, Yan, N. | | Deposit date: | 2018-08-11 | | Release date: | 2018-10-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta 1.
Science, 362, 2018
|
|
7X45
 
 | | Grass carp interferon gamma related | | Descriptor: | Interferon gamma | | Authors: | Wang, J, Zou, J, Zhu, X. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Novel Dimeric Architecture of an IFN-gamma-Related Cytokine Provides Insights into Subfunctionalization of Type II IFNs in Teleost Fish.
J Immunol., 209, 2022
|
|
5WYK
 
 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Mtr4-depleted, Enp1-TAP) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2017-05-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5WY3
 
 | |
7LQ7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV503 and COVA1-16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 13, 2021
|
|
7EGR
 
 | | Co-crystal structure of Ac-AChBPP in complex with RgIA | | Descriptor: | MAGNESIUM ION, RgIA, Soluble acetylcholine receptor | | Authors: | Wang, X.Q, Pan, S, Fan, Y.X, Xue, Y, Zhu, X.P, Luo, S.L. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Co-crystal structure of Ac-AChBPP in complex with RgIA
To Be Published
|
|
