7ECF
 
 | | Crystal Structure of d(G4C2)2-Ba in C2221 space group | | Descriptor: | BARIUM ION, DNA (5'-D(*GP*GP*GP*GP*CP*CP*GP*GP*GP*GP*CP*C)-3') | | Authors: | Geng, Y, Liu, C, Cai, Q, Luo, Z, Zhu, G. | | Deposit date: | 2021-03-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of parallel G-quadruplex formed by the two-repeat ALS- and FTD-related GGGGCC sequence.
Nucleic Acids Res., 49, 2021
|
|
7ECG
 
 | | Crystal Structure of d(G4C2)2-Ba in F222 space group | | Descriptor: | BARIUM ION, DNA (5'-D(*GP*GP*GP*GP*CP*CP*GP*GP*GP*GP*CP*C)-3') | | Authors: | Geng, Y, Liu, C, Cai, Q, Luo, Z, Zhu, G. | | Deposit date: | 2021-03-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of parallel G-quadruplex formed by the two-repeat ALS- and FTD-related GGGGCC sequence.
Nucleic Acids Res., 49, 2021
|
|
6WO0
 
 | | human Artemis/SNM1C catalytic domain, crystal form 1 | | Descriptor: | GLYCEROL, Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
6WNL
 
 | | human Artemis/SNM1C catalytic domain, crystal form 2 | | Descriptor: | Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
4LYO
 
 | | CROSS-LINKED CHICKEN LYSOZYME CRYSTAL IN NEAT ACETONITRILE, THEN BACK-SOAKED IN WATER | | Descriptor: | LYSOZYME | | Authors: | Huang, Q, Wang, Z, Zhu, G, Qian, M, Shao, M, Jia, Y, Tang, Y. | | Deposit date: | 1998-03-11 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray studies on cross-linked lysozyme crystals in acetonitrile-water mixture.
Biochim.Biophys.Acta, 1384, 1998
|
|
4MYS
 
 | | 1.4 Angstrom Crystal Structure of 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase with SHCHC and Pyruvate | | Descriptor: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, GLYCEROL, ... | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-28 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
8X0S
 
 | | Crystal structure of r(G4C2)2 | | Descriptor: | POTASSIUM ION, RNA (5'-R(*GP*GP*GP*GP*CP*CP*GP*GP*GP*GP*C)-3') | | Authors: | Geng, Y, Liu, C, Cai, Q, Zhu, G. | | Deposit date: | 2023-11-05 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Crystal structure of a tetrameric RNA G-quadruplex formed by hexanucleotide repeat expansions of C9orf72 in ALS/FTD.
Nucleic Acids Res., 52, 2024
|
|
4HHR
 
 | | Crystal Structure of fatty acid alpha-dioxygenase (Arabidopsis thaliana) | | Descriptor: | Alpha-dioxygenase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Goulah, C.C, Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2012-10-10 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The crystal structure of alpha-Dioxygenase provides insight into diversity in the cyclooxygenase-peroxidase superfamily.
Biochemistry, 52, 2013
|
|
4HHS
 
 | | Crystal Structure of fatty acid alpha-dioxygenase (Arabidopsis thaliana) | | Descriptor: | Alpha-dioxygenase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Goulah, C.C, Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2012-10-10 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of alpha-Dioxygenase provides insight into diversity in the cyclooxygenase-peroxidase superfamily.
Biochemistry, 52, 2013
|
|
4IL3
 
 | | Crystal Structure of S. mikatae Ste24p | | Descriptor: | Ste24p, ZINC ION | | Authors: | Pryor Jr, E.E, Horanyi, P.S, Clark, K, Fedoriw, N, Connelly, S.M, Koszelak-Rosenblum, M, Zhu, G, Malkowski, M.G, Dumont, M.E, Wiener, M.C, Membrane Protein Structural Biology Consortium (MPSBC) | | Deposit date: | 2012-12-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structure of the integral membrane protein CAAX protease Ste24p.
Science, 339, 2013
|
|
8HT7
 
 | | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*TP*GP*GP*G)-3'), GLN-ALA-GLN-ALA-THR-ILE-SER-PHE-PRO-LYS-ARG-LYS-LEU-SER-TRP | | Authors: | Liu, C, Zhu, G, Geng, Y, Xu, N. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex.
Int.J.Biol.Macromol., 260, 2024
|
|
5YEY
 
 | |
7MF0
 
 | | Co-crystal structure of PERK with inhibitor (R)-2-amino-N-cyclopropyl-5-(4-(2-(3,5-difluorophenyl)-2-hydroxyacetamido)-2-methylphenyl)nicotinamide | | Descriptor: | 2-amino-N-cyclopropyl-5-(4-{[(2R)-2-(3,5-difluorophenyl)-2-hydroxyacetyl]amino}-2-methylphenyl)pyridine-3-carboxamide, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wiens, B, Koszelak-Rosenblum, M, Surman, M.D, Zhu, G, Mulvihill, M.J. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Discovery of 2-amino-3-amido-5-aryl-pyridines as highly potent, orally bioavailable, and efficacious PERK kinase inhibitors.
Bioorg.Med.Chem.Lett., 43, 2021
|
|
2TIO
 
 | | LOW PACKING DENSITY FORM OF BOVINE BETA-TRYPSIN IN CYCLOHEXANE | | Descriptor: | BENZAMIDINE, CALCIUM ION, HEXANE, ... | | Authors: | Huang, Q, Zhu, G, Tang, Q. | | Deposit date: | 1998-09-23 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray studies on two forms of bovine beta-trypsin crystals in neat cyclohexane.
Biochim.Biophys.Acta, 1429, 1998
|
|
2W1O
 
 | | NMR structure of dimerization domain of human ribosomal protein P2 | | Descriptor: | 60S ACIDIC RIBOSOMAL PROTEIN P2 | | Authors: | Lee, K.M, Chan, D.S, Sze, K.H, Zhu, G, Shaw, P.C, Wong, K.B. | | Deposit date: | 2008-10-20 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Dimerization Domain of Ribosomal Protein P2 Provides Insights for the Structural Organization of Eukaryotic Stalk.
Nucleic Acids Res., 38, 2010
|
|
2VS6
 
 | | K173A, R174A, K177A-trichosanthin | | Descriptor: | RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Too, P.H, Ma, M.K, Mak, A.N, Tung, C.K, Zhu, G, Au, S.W, Wong, K.B, Shaw, P.C, Ng, A. | | Deposit date: | 2008-04-21 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Nucleic Acids Res., 37, 2009
|
|
6JKN
 
 | | Crystal Structure of G-quadruplex Formed by Bromo-substituted Human Telomeric DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*AP*GP*(BGM)P*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*(BGM)P*G)-3'), POTASSIUM ION | | Authors: | Geng, Y, Cai, Q, Liu, C, Zhu, G. | | Deposit date: | 2019-03-01 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | The crystal structure of an antiparallel chair-type G-quadruplex formed by Bromo-substituted human telomeric DNA.
Nucleic Acids Res., 47, 2019
|
|
6KVG
 
 | | The solution structure of human Orc6 | | Descriptor: | Origin recognition complex subunit 6 | | Authors: | Liu, C, Xu, N, You, Y, Zhu, G. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA replication origin recognition by human Orc6 protein binding with DNA.
Nucleic Acids Res., 48, 2020
|
|
7W68
 
 | | human single hexameric Mcm2-7 complex | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Xu, N.N, Lin, Q.P, Liu, C.D, Tian, H.L, Xiang, Y, Zhu, G. | | Deposit date: | 2021-12-01 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | human single hexameric Mcm2-7 complex
To Be Published
|
|
2KNO
 
 | |
2JJR
 
 | | V232K, N236D-trichosanthin | | Descriptor: | DI(HYDROXYETHYL)ETHER, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN, SULFATE ION, ... | | Authors: | Too, P.H, Ma, M.K, Mak, A.N, Tung, C.K, Zhu, G, Au, S.W, Wong, K.B, Shaw, P.C. | | Deposit date: | 2008-04-21 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Nucleic Acids Res., 37, 2009
|
|
2JDL
 
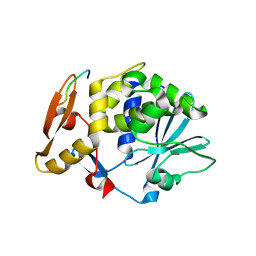 | | Structure of C-terminal region of acidic P2 ribosomal protein complexed with trichosanthin | | Descriptor: | ACIDIC RIBOSOMAL PROTEIN P2, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Too, P.H, Mak, A.N, Zhu, G, Au, S.W, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Nucleic Acids Res., 37, 2009
|
|
2Q12
 
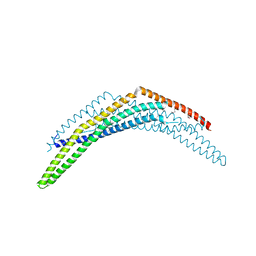 | | Crystal Structure of BAR domain of APPL1 | | Descriptor: | DCC-interacting protein 13 alpha | | Authors: | Zhang, X.C, Zhu, G. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5.
Embo J., 26, 2007
|
|
2LE8
 
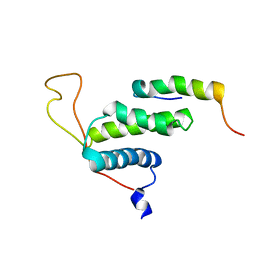 | | The protein complex for DNA replication | | Descriptor: | DNA replication factor Cdt1, DNA replication licensing factor MCM6 | | Authors: | Liu, C, Wei, Z, Zhu, G. | | Deposit date: | 2011-06-14 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the Cdt1-mediated MCM2-7 chromatin loading
Nucleic Acids Res., 40, 2012
|
|
2LOZ
 
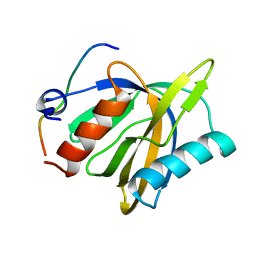 | | The novel binding mode of DLC1 and Tensin2 PTB domain | | Descriptor: | Rho GTPase-activating protein 7, Tensin-like C1 domain-containing phosphatase | | Authors: | Liu, C, Chen, L, Zhu, G. | | Deposit date: | 2012-01-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the phosphotyrosine binding (PTB) domain of human tensin2 protein in complex with deleted in liver cancer 1 (DLC1) peptide reveals a novel peptide binding mode.
J.Biol.Chem., 287, 2012
|
|
