4IH1
 
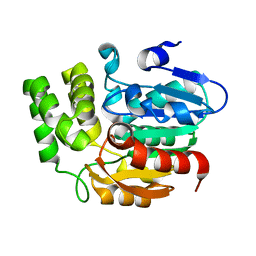 | | Crystal structure of Karrikin Insensitive 2 (KAI2) from Arabidopsis thaliana | | Descriptor: | Hydrolase, alpha/beta fold family protein | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH9
 
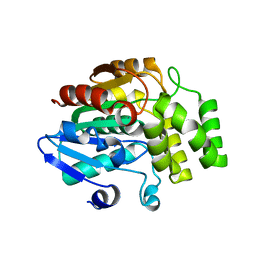 | | Crystal structure of rice DWARF14 (D14) | | Descriptor: | Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IHA
 
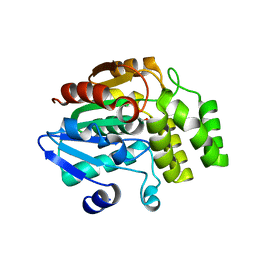 | | Crystal structure of rice DWARF14 (D14) in complex with a GR24 hydrolysis intermediate | | Descriptor: | (2R,3R)-2,4,4-trihydroxy-3-methylbutanal, Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH4
 
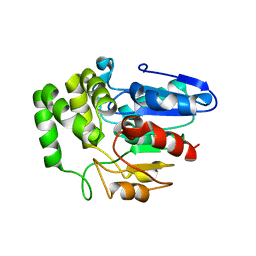 | | Crystal structure of Arabidopsis DWARF14 orthologue, AtD14 | | Descriptor: | AT3g03990/T11I18_10 | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
8J3A
 
 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
Y54C mutant in complex with PF00835231
To Be Published
|
|
8J37
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
K90R mutant in complex with PF00835231
To Be Published
|
|
8J35
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF00835231
To Be Published
|
|
8J32
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231
To Be Published
|
|
8J38
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
P132H mutant in complex with PF00835231
To Be Published
|
|
8J3B
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF00835231
To Be Published
|
|
3GYU
 
 | | Nuclear receptor DAF-12 from parasitic nematode Strongyloides stercoralis in complex with its physiological ligand dafachronic acid delta 7 | | Descriptor: | (5beta,14beta,17alpha,25R)-3-oxocholest-7-en-26-oic acid, Nuclear hormone receptor of the steroid/thyroid hormone receptors superfamily, SRC1 | | Authors: | Zhou, X.E, Wang, Z, Suino-Powell, K, Motola, D.L, Conneely, A, Ogata, C, Sharma, K.K, Auchus, R.J, Kliewer, S.A, Xu, H.E, Mangelsdorf, D.J. | | Deposit date: | 2009-04-05 | | Release date: | 2009-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8J39
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with PF00835231
To Be Published
|
|
8J36
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
M49I mutant in complex with PF00835231
To Be Published
|
|
3GYT
 
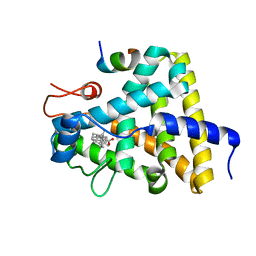 | | Nuclear receptor DAF-12 from parasitic nematode Strongyloides stercoralis in complex with its physiological ligand dafachronic acid delta 4 | | Descriptor: | (14beta,17alpha,25R)-3-oxocholest-4-en-26-oic acid, Nuclear hormone receptor of the steroid/thyroid hormone receptors superfamily, SRC1 | | Authors: | Zhou, X.E, Wang, Z, Suino-Powell, K, Motola, D.L, Conneely, A, Ogata, C, Sharma, K.K, Auchus, R.J, Kliewer, S.A, Xu, H.E, Mangelsdorf, D.J. | | Deposit date: | 2009-04-05 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8J34
 
 | | Crystal structure of MERS main protease in complex with PF00835231 | | Descriptor: | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of MERS main protease in complex with PF00835231
To Be Published
|
|
3P0U
 
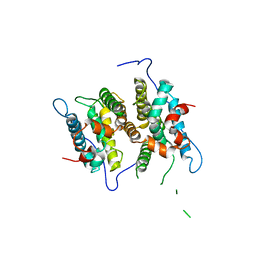 | | Crystal Structure of the ligand binding domain of human testicular receptor 4 | | Descriptor: | Nuclear receptor subfamily 2 group C member 2 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Xu, Y, Chan, C.-W, Kruse, S.W, Reynolds, R, Engel, J.D, Xu, H.E. | | Deposit date: | 2010-09-29 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Orphan Nuclear Receptor TR4 Is a Vitamin A-activated Nuclear Receptor.
J.Biol.Chem., 286, 2011
|
|
4RED
 
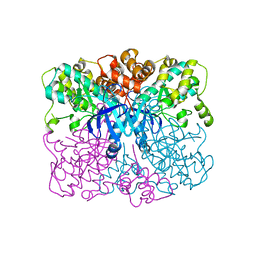 | | Crystal structure of human AMPK alpha1 KD-AID with K43A mutation | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
4RER
 
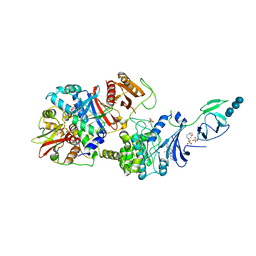 | | Crystal structure of the phosphorylated human alpha1 beta2 gamma1 holo-AMPK complex bound to AMP and cyclodextrin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, ... | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.047 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
4REW
 
 | | Crystal structure of the non-phosphorylated human alpha1 beta2 gamma1 holo-AMPK complex | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-24 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.58 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
8EUP
 
 | | Ytm1 associated 60S nascent ribosome State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUG
 
 | | Ytm1 associated nascent 60S ribosome State 3 | | Descriptor: | 60S ribosomal protein L10-A, 60S ribosomal protein L11-A, 60S ribosomal protein L13, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EV3
 
 | | Ytm1 associated 60S nascent ribosome (-Fkbp39) State 1B | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETJ
 
 | | Fkbp39 associated 60S nascent ribosome State 2 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ESQ
 
 | | Ytm1 associated nascent 60S ribosome State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
