7YSP
 
 | |
7YSR
 
 | | GTPgammaS MT decorated with kinesin | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Tubulin alpha chain, ... | | Authors: | Zhou, J, Wang, H.-W. | | Deposit date: | 2022-08-12 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into the mechanism of GTP initiation of microtubule assembly.
Nat Commun, 14, 2023
|
|
7YSN
 
 | | Tubulin heterodimer structure of GMPCPP state in solution | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tubulin alpha-1B chain, ... | | Authors: | Zhou, J, Wang, H.-W. | | Deposit date: | 2022-08-12 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the mechanism of GTP initiation of microtubule assembly.
Nat Commun, 14, 2023
|
|
7YSQ
 
 | | GTPgammaS Tube decorated with kinesin | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Tubulin alpha chain, ... | | Authors: | Zhou, J, Wang, H.-W. | | Deposit date: | 2022-08-12 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural insights into the mechanism of GTP initiation of microtubule assembly.
Nat Commun, 14, 2023
|
|
7YSO
 
 | |
6LG3
 
 | |
3LWV
 
 | | Structure of H/ACA RNP bound to a substrate RNA containing 2'-deoxyuridine | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(du)P*GP*CP*GP*GP*UP*UP*U)-3', 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Functional and Structural Impact of Target Uridine Substitutions on the H/ACA Ribonucleoprotein Particle Pseudouridine Synthase .
Biochemistry, 49, 2010
|
|
3LWO
 
 | | Structure of H/ACA RNP bound to a substrate RNA containing 5BrU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(5BU)P*GP*CP*GP*GP*UP*UP*U)-3', 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.855 Å) | | Cite: | Glycosidic bond conformation preference plays a pivotal role in catalysis of RNA pseudouridylation: a combined simulation and structural study.
J.Mol.Biol., 401, 2010
|
|
3LWP
 
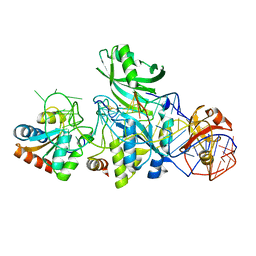 | | Structure of H/ACA RNP bound to a substrate RNA containing 5BrdU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(BRU)P*GP*CP*GP*GP*UP*UP*U)-3, 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glycosidic bond conformation preference plays a pivotal role in catalysis of RNA pseudouridylation: a combined simulation and structural study.
J.Mol.Biol., 401, 2010
|
|
3LWR
 
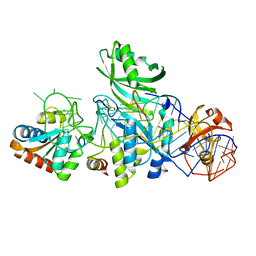 | | Structure of H/ACA RNP bound to a substrate RNA containing 4SU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(4SU)P*GP*CP*GP*GP*UP*UP*U)-3', H/ACA RNA, Large ribosomal subunit protein eL8, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Functional and Structural Impact of Target Uridine Substitutions on the H/ACA Ribonucleoprotein Particle Pseudouridine Synthase .
Biochemistry, 49, 2010
|
|
3MQK
 
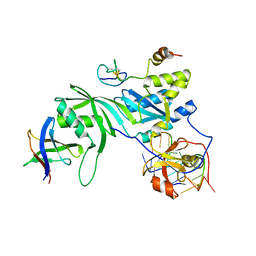 | | Cbf5-Nop10-Gar1 complex binding with 17mer RNA containing ACA trinucleotide | | Descriptor: | RNA (5'-R(P*CP*GP*AP*UP*CP*CP*AP*CP*A)-3'), RNA (5'-R(P*GP*UP*UP*CP*GP*AP*UP*CP*CP*AP*CP*AP*G)-3'), Ribosome biogenesis protein Nop10, ... | | Authors: | Zhou, J, Liang, B, Li, H. | | Deposit date: | 2010-04-28 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional evidence of high specificity of Cbf5 for ACA trinucleotide.
Rna, 17, 2011
|
|
3LWQ
 
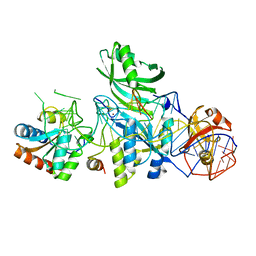 | | Structure of H/ACA RNP bound to a substrate RNA containing 3MU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(UR3)P*GP*CP*GP*GP*UP*UP*U)-3, 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.678 Å) | | Cite: | Functional and Structural Impact of Target Uridine Substitutions on the H/ACA Ribonucleoprotein Particle Pseudouridine Synthase .
Biochemistry, 49, 2010
|
|
8VPG
 
 | |
8VPI
 
 | |
8VPH
 
 | |
8HS0
 
 | | The mutant structure of DHAD V178W | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural bases of dihydroxy acid dehydratase inhibition and biodesign for self-resistance
Biodes Res, 0, 2024
|
|
8IKZ
 
 | | The mutant structure of DHAD | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural bases of dihydroxy acid dehydratase inhibition and biodesign for self-resistance
Biodes Res, 0, 2024
|
|
8IMU
 
 | | Dihydroxyacid dehydratase (DHAD) mutant-V497F | | Descriptor: | ACETATE ION, Dihydroxy-acid dehydratase, chloroplastic, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural bases of dihydroxy acid dehydratase inhibition and biodesign for self-resistance
Biodes Res, 0, 2024
|
|
8CY5
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 39 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-{3-[1-(tert-butoxycarbonyl)piperidin-4-yl]propyl}adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY3
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-[3-(4-aminophenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY1
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 19 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(5-phenylpentyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXZ
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-(3-Phenylpropyl)adenosine (Compound 14) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(3-phenylpropyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY4
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 16 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-[3-(4-hydroxyphenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY0
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor MC4756 (Compound 178) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(4-phenylbutyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
6KAB
 
 | |
