8HLQ
 
 | |
8HKN
 
 | |
7FCJ
 
 | | Zebrafish SIGIRR TIR domain mutant - C299S | | Descriptor: | SIGIRR protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FCH
 
 | | IL-18Rbeta TIR domain | | Descriptor: | Interleukin-18 receptor accessory protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FCL
 
 | | Zebrafish SIGIRR TIR domain | | Descriptor: | SIGIRR protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
5WRQ
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 5-[[2,4-bis(oxidanylidene)quinazolin-1-yl]methyl]-2-fluoranyl-N-[(3R)-1-(3-methylbutyl)pyrrolidin-3-yl]benzamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-03 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5WS1
 
 | | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor | | Descriptor: | 2-[(3R)-3-azanylpyrrolidin-1-yl]carbonyl-1H-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor
To Be Published
|
|
5WTC
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-[4-[2,2,2-tris(fluoranyl)ethyl]piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5WS0
 
 | | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor | | Descriptor: | 2-piperazin-1-ylcarbonyl-1H-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor
To Be Published
|
|
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | Descriptor: | D-Cysteine desulfhydrase | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | Descriptor: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7VA1
 
 | |
7EGV
 
 | | Acetolactate Synthase from Trichoderma harzianum with inhibitor harzianic acid | | Descriptor: | (2S)-3-methyl-2-[[(2S,4R)-1-methyl-4-[(2E,4E)-octa-2,4-dienoyl]-3,5-bis(oxidanylidene)pyrrolidin-2-yl]methyl]-2-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Zang, X, Xie, L, Chen, M, Tang, Y, Zhou, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Harzianic Acid from Trichoderma afroharzianum Is a Natural Product Inhibitor of Acetohydroxyacid Synthase.
J.Am.Chem.Soc., 2021
|
|
7EHE
 
 | | Acetolactate Synthase from Trichoderma harzianum | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Acetolactate synthase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zang, X, Tang, Y, Zhou, J. | | Deposit date: | 2021-03-29 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Harzianic Acid from Trichoderma afroharzianum Is a Natural Product Inhibitor of Acetohydroxyacid Synthase.
J.Am.Chem.Soc., 2021
|
|
7EEY
 
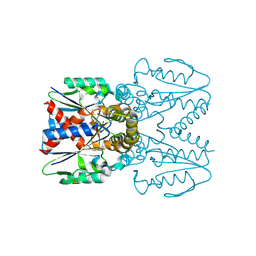 | |
7EZP
 
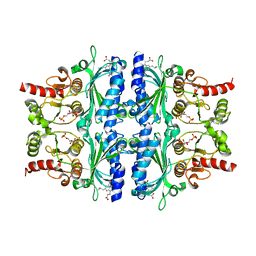 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
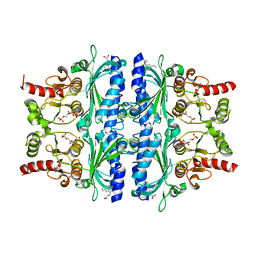 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7XP9
 
 | |
7XPC
 
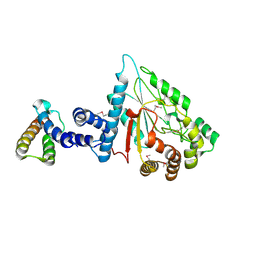 | |
6LCV
 
 | |
6LCU
 
 | |
