8UF3
 
 | |
8Z36
 
 | | Crystal structure of HOIP PUB domain in complex with sertraline complex | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, E3 ubiquitin-protein ligase RNF31 | | Authors: | Zhong, F, Ruan, K. | | Deposit date: | 2024-04-14 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Repurposing Tolfenamic Acid to Anchor the Uncharacterized Pocket of the PUB Domain for Proteolysis of the Atypical E3 Ligase HOIP.
Acs Chem.Biol., 19, 2024
|
|
8Z30
 
 | | Crystal structure of HOIP PUB domain in complex with tolfenamic acid complex | | Descriptor: | 2-[(3-chloro-2-methylphenyl)amino]benzoic acid, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Zhong, F, Ruan, K. | | Deposit date: | 2024-04-14 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Repurposing Tolfenamic Acid to Anchor the Uncharacterized Pocket of the PUB Domain for Proteolysis of the Atypical E3 Ligase HOIP.
Acs Chem.Biol., 19, 2024
|
|
7WQH
 
 | | Crystal structure of HCoV-NL63 main protease with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhong, F.L, Zhou, X.L, Lin, C, Zeng, P, Li, J, Zhang, J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7DR9
 
 | |
8ZF1
 
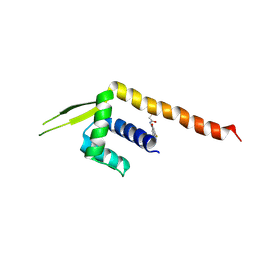 | | Crystal structure of a Chemo Triplet Photoenzyme (CTPe) | | Descriptor: | N-(9-oxidanylidenethioxanthen-2-yl)ethanamide, Transcriptional regulator, PadR-like family | | Authors: | Chen, X, Qian, J.Y, Guo, J, Wu, Y.Z, Zhong, F.R. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chemogenetic Evolution of Diversified Photoenzymes for Enantioselective [2 + 2] Cycloadditions in Whole Cells.
J.Am.Chem.Soc., 146, 2024
|
|
9IMB
 
 | |
6Q2U
 
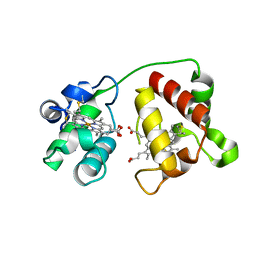 | | Structure of Cytochrome C4 from Pseudomonas aeruginosa | | Descriptor: | Cytochrome c4, HEME C | | Authors: | Carpenter, J.M, Zhong, F, Pletneva, E.V, Ragusa, M.J. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and redox properties of the diheme electron carrier cytochrome c4from Pseudomonas aeruginosa.
J.Inorg.Biochem., 203, 2019
|
|
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
7C2Q
 
 | | The crystal structure of COVID-19 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Hu, X.H, Zhou, H, Wang, Q.S, Li, j, Zhang, J. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of SARS-CoV-2 main protease in the apo state.
Sci China Life Sci, 64, 2021
|
|
7C2Y
 
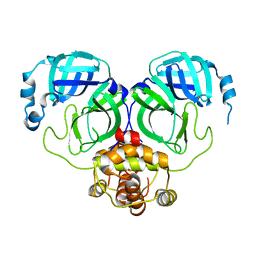 | | The crystal structure of COVID-2019 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhou, H, Hu, X.H, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-05-10 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | COVID-2019 main protease in the apo state
To Be Published
|
|
4Q5P
 
 | | Lysine-Ligated Yeast Iso-1 Cytochrome C | | Descriptor: | Cytochrome c iso-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Amacher, J.F, Zhu, M.Q, Zhong, F, Pletneva, E.V, Madden, D.R. | | Deposit date: | 2014-04-17 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A Compact Structure of Cytochrome c Trapped in a Lysine-Ligated State: Loop Refolding and Functional Implications of a Conformational Switch.
J.Am.Chem.Soc., 137, 2015
|
|
7VVP
 
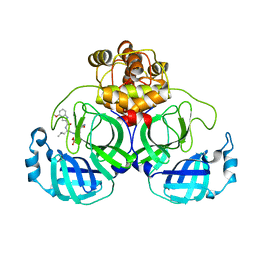 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7VLQ
 
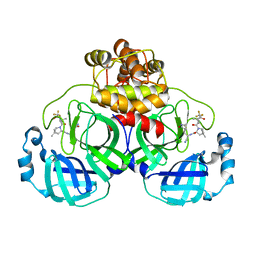 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P212121 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.939106 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLO
 
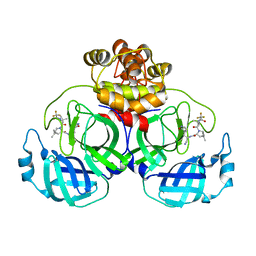 | | Crystal structure of SARS coronavirus main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.0227 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P1211 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.50251937 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VTC
 
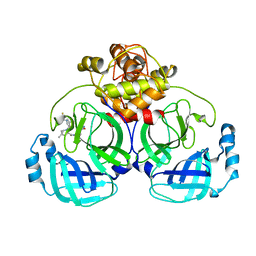 | | Crystal structure of MERS main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53865623 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7XUP
 
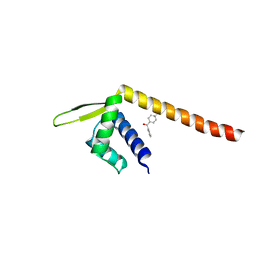 | | Crystal structure of TPe3.0 | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
7XUQ
 
 | | Crystal structure of Tpe3.0 complexed with N-Boc-3-alkenylindole | | Descriptor: | Transcriptional regulator, PadR-like family, dimethyl 2-[[2-methyl-1-[(2-methylpropan-2-yl)oxycarbonyl]indol-3-yl]methyl]-2-prop-2-enyl-propanedioate | | Authors: | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7E6R
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.6 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-23 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6L
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.0 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78037143 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6M
 
 | | Crystal structure of Human coronavirus NL63 3C-like protease | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83445024 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6N
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.2 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8413 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
