4USG
 
 | | Crystal structure of PC4 W89Y mutant complex with DNA | | 分子名称: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*TP*G)-3', ACTIVATED RNA POLYMERASE II TRANSCRIPTIONAL COACTIVATOR P15 | | 著者 | Zhao, Y, Liu, J. | | 登録日 | 2014-07-08 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.973 Å) | | 主引用文献 | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
4V0R
 
 | | DENGUE VIRUS FULL LENGTH NS5 COMPLEXED WITH GTP AND SAH | | 分子名称: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Zhao, Y, Soh, S, Zheng, J, Phoo, W.W, Swaminathan, K, Cornvik, T.C, Lim, S.P, Shi, P.-Y, Lescar, J, Vasudevan, S.G, Luo, D. | | 登録日 | 2014-09-18 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A Crystal Structure of the Dengue Virus Ns5 Protein Reveals a Novel Inter-Domain Interface Essential for Protein Flexibility and Virus Replication.
Plos Pathog., 11, 2015
|
|
5IZ8
 
 | | Protein-protein interaction | | 分子名称: | ACE-ALA-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein, TRIETHYLENE GLYCOL | | 著者 | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | 登録日 | 2016-03-25 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZA
 
 | | Protein-protein interaction | | 分子名称: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | 著者 | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | 登録日 | 2016-03-25 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
1QQC
 
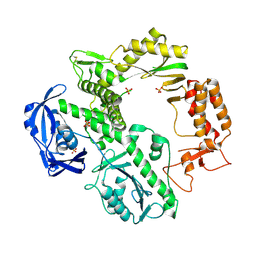 | | CRYSTAL STRUCTURE OF AN ARCHAEBACTERIAL DNA POLYMERASE D.TOK | | 分子名称: | DNA POLYMERASE II, MAGNESIUM ION, SULFATE ION | | 著者 | Zhao, Y, Jeruzalmi, D, Leighton, L, Lasken, R, Kuriyan, J. | | 登録日 | 1999-06-02 | | 公開日 | 1999-10-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of an archaebacterial DNA polymerase
Structure Fold.Des., 7, 1999
|
|
7R9H
 
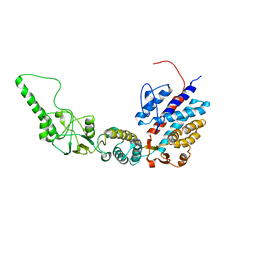 | | Methanococcus maripaludis chaperonin, open conformation 2 | | 分子名称: | Chaperonin | | 著者 | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | 登録日 | 2021-06-29 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9M
 
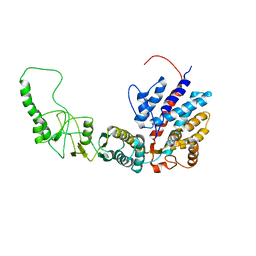 | | Methanococcus maripaludis chaperonin, closed conformation 2 | | 分子名称: | Chaperonin | | 著者 | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | 登録日 | 2021-06-29 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9J
 
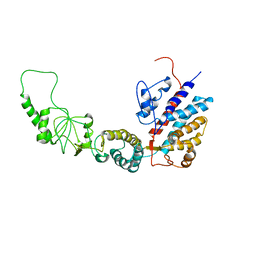 | | Methanococcus maripaludis chaperonin, open conformation 4 | | 分子名称: | Chaperonin | | 著者 | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | 登録日 | 2021-06-29 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9I
 
 | | Methanococcus maripaludis chaperonin, open conformation 2 | | 分子名称: | Chaperonin | | 著者 | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | 登録日 | 2021-06-29 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9O
 
 | | Methanococcus maripaludis chaperonin, closed conformation 1 | | 分子名称: | Chaperonin | | 著者 | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | 登録日 | 2021-06-29 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9K
 
 | | Methanococcus maripaludis chaperonin, closed conformation 4 | | 分子名称: | Chaperonin | | 著者 | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | 登録日 | 2021-06-29 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9U
 
 | | Methanococcus maripaludis chaperonin, closed conformation 3 | | 分子名称: | Chaperonin | | 著者 | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | 登録日 | 2021-06-29 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9E
 
 | | Methanococcus maripaludis chaperonin, open conformation 1 | | 分子名称: | Chaperonin | | 著者 | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | 登録日 | 2021-06-29 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7RAK
 
 | | Methanococcus maripaludis chaperonin complex in open conformation | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin | | 著者 | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | 登録日 | 2021-07-01 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
1R5I
 
 | | Crystal structure of the MAM-MHC complex | | 分子名称: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | 著者 | Zhao, Y, Li, Z, Drozd, S.J, Guo, Y, Mourad, W, Li, H. | | 登録日 | 2003-10-10 | | 公開日 | 2004-03-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Mycoplasma arthritidis mitogen complexed with HLA-DR1 reveals a novel superantigen fold and a dimerized superantigen-MHC complex.
Structure, 12, 2004
|
|
7X79
 
 | | The crystal structure of human Calpain-1 protease core in complex with 14b | | 分子名称: | CALCIUM ION, Calpain-1 catalytic subunit, HYDROSULFURIC ACID, ... | | 著者 | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | 登録日 | 2022-03-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of human Calpain-1 protease core in complex with 14a
To Be Published
|
|
2Q6N
 
 | | Structure of Cytochrome P450 2B4 with Bound 1-(4-cholorophenyl)imidazole | | 分子名称: | 1-(4-CHLOROPHENYL)-1H-IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Zhao, Y, Sun, L, Muralidhara, B.K, Kumar, S, White, M.A, Stout, C.D, Halpert, J.R. | | 登録日 | 2007-06-05 | | 公開日 | 2007-11-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4.
Biochemistry, 46, 2007
|
|
6LPM
 
 | |
7XF3
 
 | | The structure of HLA-B*1501/BM58-66AF9 | | 分子名称: | 9-mer peptide from Matrix protein 1, Beta-2-microglobulin, MHC class I antigen | | 著者 | Zhao, Y.Z, Xiao, W.L, Wu, Y.N, Fan, W.F, Yue, C, Zhang, Q.X, Zhang, D.N, Yuan, X.J, Yao, S.J, Liu, S, Li, M, Wang, P.Y, Zhang, H.J, Zhang, J, Zhao, M, Zheng, X.Q, Liu, W.J, Gao, G.F, Liu, W.L. | | 登録日 | 2022-03-31 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Parallel T Cell Immunogenic Regions in Influenza B and A Viruses with Distinct Nuclear Export Signal Functions: The Balance between Viral Life Cycle and Immune Escape.
J Immunol., 210, 2023
|
|
8JGR
 
 | |
8JGT
 
 | |
8JGX
 
 | |
8JGW
 
 | |
8JGU
 
 | |
8JGQ
 
 | |
