2Q2G
 
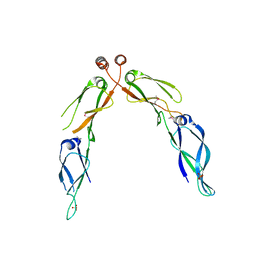 | | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800 | | Descriptor: | Heat shock 40 kDa protein, putative (fragment), SULFATE ION | | Authors: | Wernimont, A.K, Lew, J, Lin, L, Hassanali, A, Kozieradzki, I, Wasney, G, Vedadi, M, Walker, J.R, Zhao, Y, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800.
To be Published
|
|
2B0R
 
 | | Crystal Structure of Cyclase-Associated Protein from Cryptosporidium parvum | | Descriptor: | UNKNOWN ATOM OR ION, possible adenyl cyclase-associated protein | | Authors: | Tempel, W, Dong, A, Zhao, Y, Lew, J, Kozieradzki, I, Alam, Z, Melone, M, Wasney, G, Vedadi, M, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Hui, R, Bochkarev, A, Artz, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of a G-actin sequestering protein with a vital role in malaria oocyst development inside the mosquito vector
J.Biol.Chem., 151, 2010
|
|
6L2W
 
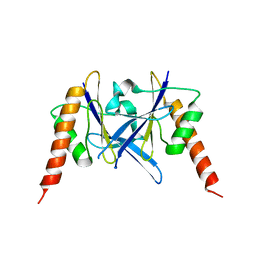 | | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1 | | Descriptor: | freshwater cyanophage protein | | Authors: | Wang, Y, Jin, H, Yang, F, Jiang, Y.L, Zhao, Y.Y, Chen, Z.P, Li, W.F, Chen, Y, Zhou, C.Z, Li, Q. | | Deposit date: | 2019-10-07 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1.
Proteins, 88, 2020
|
|
4ZOW
 
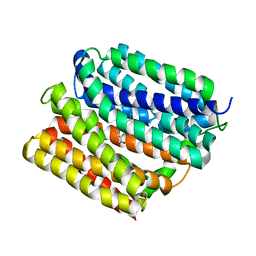 | | Crystal structure of E. coli multidrug transporter MdfA in complex with chloramphenicol | | Descriptor: | CHLORAMPHENICOL, Multidrug transporter MdfA | | Authors: | Zhang, X.C, Heng, J, Zhao, Y, Wang, X. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate-bound structure of the E. coli multidrug resistance transporter MdfA
Cell Res., 25, 2015
|
|
4ZP2
 
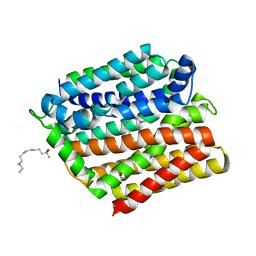 | | Crystal structure of E. coli multidrug transporter MdfA in complex with n-dodecyl-N,N-dimethylamine-N-oxide | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Multidrug transporter MdfA | | Authors: | Zhang, X.C, Heng, J, Zhao, Y, Wang, X. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-bound structure of the E. coli multidrug resistance transporter MdfA
Cell Res., 25, 2015
|
|
4ZP0
 
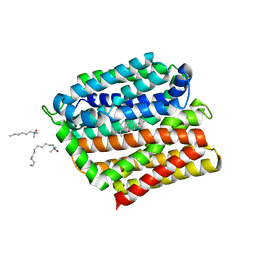 | | Crystal structure of E. coli multidrug transporter MdfA in complex with deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, LAURYL DIMETHYLAMINE-N-OXIDE, Multidrug transporter MdfA | | Authors: | Zhang, X.C, Heng, J, Zhao, Y, Wang, X. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate-bound structure of the E. coli multidrug resistance transporter MdfA
Cell Res., 25, 2015
|
|
2POY
 
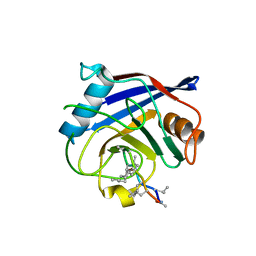 | | Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120 in complex with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryptosporidium Parvum Cyclophilin Type Peptidyl-Prolyl Cis-Trans Isomerase Cgd2_4120 in Complex with Cyclosporin A.
To be Published
|
|
2PLU
 
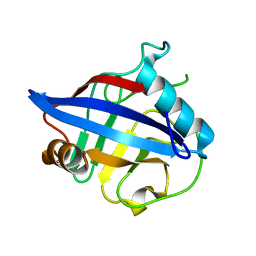 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120 | | Descriptor: | 20k cyclophilin, putative | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120.
To be Published
|
|
7LL8
 
 | | D-Protein RFX-V1 Bound to the VEGFR1 Domain 2 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V1 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
7LL9
 
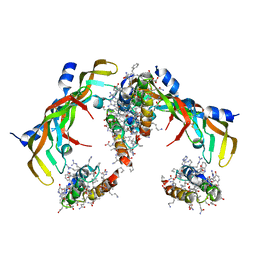 | | D-Protein RFX-V2 Bound to the VEGFR1 Domain 3 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V2 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
7WXZ
 
 | | Crystal structure of the recombinant protein HR121 from the S2 protein of SARS-CoV-2 | | Descriptor: | Spike protein S2' | | Authors: | Zheng, Y.T, Ouyang, S, Pang, W, Lu, Y, Zhao, Y.B. | | Deposit date: | 2022-02-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A variant-proof SARS-CoV-2 vaccine targeting HR1 domain in S2 subunit of spike protein.
Cell Res., 32, 2022
|
|
3CC9
 
 | | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Putative farnesyl pyrophosphate synthase, SODIUM ION | | Authors: | Wernimont, A.K, Dunford, J, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate.
To be Published
|
|
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
4ZEL
 
 | | Human dopamine beta-hydroxylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Vendelboe, T.V, Harris, P, Christensen, H.E.M, Harlos, K, Walter, T, Zhao, Y, Omari, K. | | Deposit date: | 2015-04-20 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of human dopamine beta-hydroxylase at 2.9 angstrom resolution.
Sci Adv, 2, 2016
|
|
3GUE
 
 | | Crystal Structure of UDP-glucose phosphorylase from Trypanosoma Brucei, (Tb10.389.0330) | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Wernimont, A.K, Marino, K, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-29 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of UDP-glucose phosphorylase from Trypanosoma Brucei, (Tb10.389.0330)
To be Published
|
|
3BYV
 
 | | Crystal structure of Toxoplasma gondii specific rhoptry antigen kinase domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Rhoptry kinase | | Authors: | Wernimont, A.K, Lunin, V.V, Yang, C, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Sibley, D, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
4Z7I
 
 | | Crystal structure of insulin regulated aminopeptidase in complex with ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DG025 transition-state analogue enzyme inhibitor, ... | | Authors: | Mpakali, A, Saridakis, E, Harlos, K, Zhao, Y, Stratikos, E. | | Deposit date: | 2015-04-07 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal Structure of Insulin-Regulated Aminopeptidase with Bound Substrate Analogue Provides Insight on Antigenic Epitope Precursor Recognition and Processing.
J Immunol., 195, 2015
|
|
3VT1
 
 | | Crystal structure of Ct1,3Gal43A in complex with galactose | | Descriptor: | Ricin B lectin, beta-D-galactopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
3VT2
 
 | | Crystal structure of Ct1,3Gal43A in complex with isopropy-beta-D-thiogalactoside | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, GLYCEROL, Ricin B lectin | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
3I3G
 
 | | Crystal Structure of Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A | | Descriptor: | N-acetyltransferase | | Authors: | Qiu, W, Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, J Ferguson, M.A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A
To be Published
|
|
3VT0
 
 | | Crystal structure of Ct1,3Gal43A in complex with lactose | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
3VSF
 
 | | Crystal structure of 1,3Gal43A, an exo-beta-1,3-Galactanase from Clostridium thermocellum | | Descriptor: | GLYCEROL, Ricin B lectin | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-04-25 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
