2IOM
 
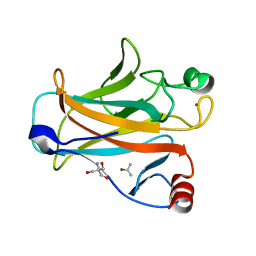 | | Mouse p53 core domain soaked with 2-propanol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular tumor antigen p53, ISOPROPYL ALCOHOL, ... | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2FQ3
 
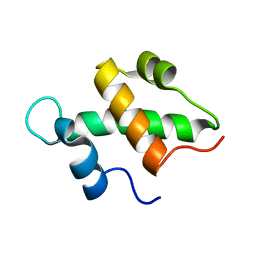 | | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes | | Descriptor: | Transcription regulatory protein SWI3 | | Authors: | Da, G, Lenkart, J, Zhao, K, Shiekhattar, R, Cairns, B.R, Marmorstein, R. | | Deposit date: | 2006-01-17 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7YR7
 
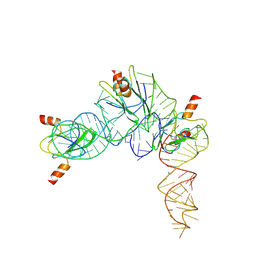 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with three RsmA protein dimers | | Descriptor: | RsmZ RNA (118-MER), Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Liu, L, Ling, X, Yang, X, Wu, Y, Liu, T, Miao, Z, Wei, X, Bujnicki, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
7YR6
 
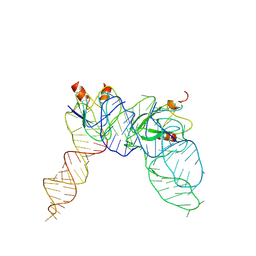 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with two RsmA protein dimers | | Descriptor: | RsmZ RNA, Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Ling, X, Yang, X, Wu, Y, Liu, T, Wei, X, Bujnick, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
5XPI
 
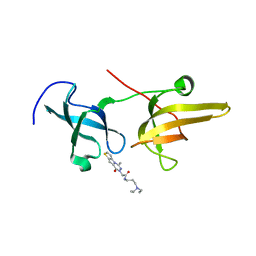 | | Structure of UHRF1 TTD in complex with NV01 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, N-[3-(diethylamino)propyl]-2-(12-methyl-9-oxidanylidene-5-thia-1,10,11-triazatricyclo[6.4.0.0^2,6]dodeca-2(6),3,7,11-tetraen-10-yl)ethanamide | | Authors: | Luo, X, Zhao, K. | | Deposit date: | 2017-06-02 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Small-Molecule Antagonists of the H3K9me3 Binding to UHRF1 Tandem Tudor Domain
SLAS Discov, 23, 2018
|
|
6AGS
 
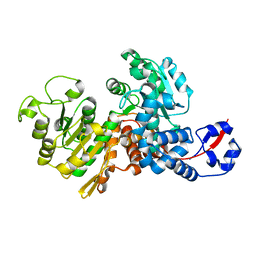 | |
7VZF
 
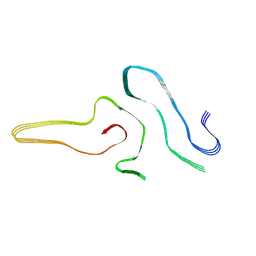 | | Cryo-EM structure of amyloid fibril formed by full-length human SOD1 | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Yuan, H.Y, Zhao, K, Zhang, M.Y, Wang, Q, Huang, X, Xu, W.C, Chen, J, Li, D, Zhang, D.L, Zou, L.Y, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human SOD1 reveals its conformational conversion.
Nat Commun, 13, 2022
|
|
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-01-18 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7EQ4
 
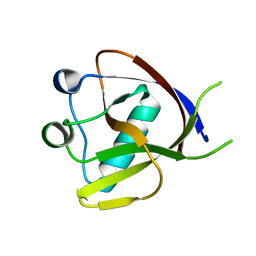 | | Crystal Structure of the N-terminus of Nonstructural protein 1 from SARS-CoV-2 | | Descriptor: | Host translation inhibitor nsp1 | | Authors: | Liu, Y, Ke, Z, Hu, H, Zhao, K, Xiao, J, Xia, Y, Li, Y. | | Deposit date: | 2021-04-29 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis and Function of the N Terminus of SARS-CoV-2 Nonstructural Protein 1.
Microbiol Spectr, 9, 2021
|
|
