8DZG
 
 | | Cryo-EM structure of bundle-forming pilus extension ATPase from E.coli in the presence of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BfpD, MAGNESIUM ION, ... | | Authors: | Nayak, A.R, Zhao, J, Donnenberg, M.S, Samso, M. | | Deposit date: | 2022-08-07 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structure of the Type IV Pilus Extension ATPase from Enteropathogenic Escherichia coli.
Mbio, 13, 2022
|
|
7L6W
 
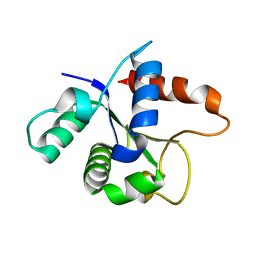 | | SFX structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Flueckiger, L, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Stacey, K.J, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
4Y2M
 
 | | apo-GolB protein | | Descriptor: | GOLD ION, Putative metal-binding transport protein | | Authors: | Wei, W, Zhao, J, Wang, F. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
7JKX
 
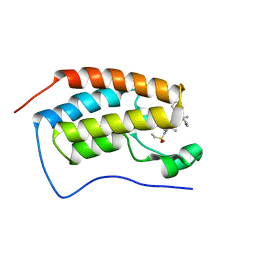 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-6 | | Descriptor: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-[1-methyl-5-(methylamino)-6-oxo-1,6-dihydropyridin-3-yl]-1H-indol-4-yl}ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-6
To Be Published
|
|
6ZT9
 
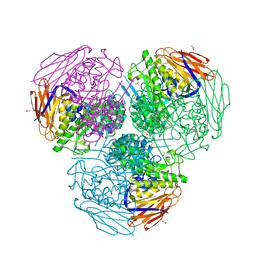 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZTA
 
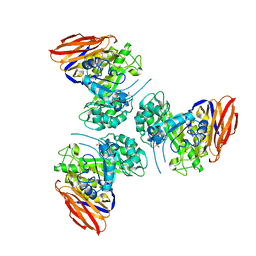 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT8
 
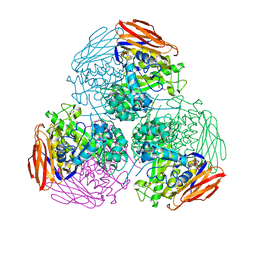 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT6
 
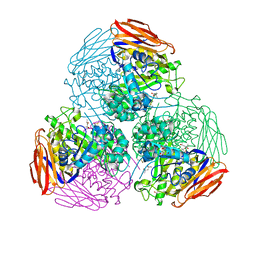 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT7
 
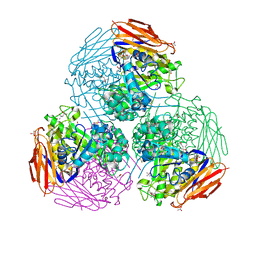 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
5KF4
 
 | |
7X79
 
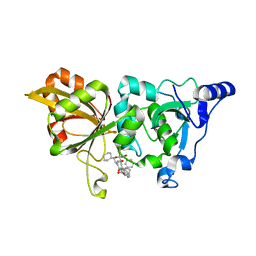 | | The crystal structure of human Calpain-1 protease core in complex with 14b | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, HYDROSULFURIC ACID, ... | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-03-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of human Calpain-1 protease core in complex with 14a
To Be Published
|
|
7BER
 
 | | SFX structure of the MyD88 TIR domain higher-order assembly (solved, rebuilt and refined using an identical protocol to the MicroED structure of the MyD88 TIR domain higher-order assembly) | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
3URM
 
 | | Crystal structure of the periplasmic sugar binding protein ChvE | | Descriptor: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-galactopyranose | | Authors: | Hu, X, Zhao, J, Binns, A, Degrado, W. | | Deposit date: | 2011-11-22 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7BEQ
 
 | | MicroED structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
3UUG
 
 | | Crystal structure of the periplasmic sugar binding protein ChvE | | Descriptor: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-glucopyranuronic acid | | Authors: | Hu, X, Zhao, J, Binns, A, Degrado, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4Y2I
 
 | | Gold ion bound to GolB | | Descriptor: | GOLD ION, Putative metal-binding transport protein | | Authors: | Wei, W, Wang, F, Ma, L, Zhao, J. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
4Y2K
 
 | | reduced form of apo-GolB | | Descriptor: | Putative metal-binding transport protein | | Authors: | Wei, W, Wang, F, Ma, L, Zhao, J. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
6QRZ
 
 | | Crystal structure of R2-like ligand-binding oxidase from Sulfolobus acidocaldarius solved by 3D micro-crystal electron diffraction | | Descriptor: | FE (III) ION, MANGANESE (III) ION, Ribonucleoside-diphosphate reductase | | Authors: | Xu, H, Lebrette, H, Clabbers, M.T.B, Zhao, J, Griese, J.J, Zou, X, Hogbom, M. | | Deposit date: | 2019-02-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Solving a new R2lox protein structure by microcrystal electron diffraction.
Sci Adv, 5, 2019
|
|
4DWT
 
 | |
1LPL
 
 | | Structural Genomics of Caenorhabditis elegans: CAP-Gly domain of F53F4.3 | | Descriptor: | Hypothetical 25.4 kDa protein F53F4.3 in chromosome V | | Authors: | Li, S, Finley, J, Liu, Z.-J, Qiu, S.H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the
Cytoskeleton-associated Protein
Glycine-rich (CAP-Gly) Domain
J.Biol.Chem., 277, 2002
|
|
1JXD
 
 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
1JXF
 
 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
4O98
 
 | | Crystal structure of Pseudomonas oleovorans PoOPH mutant H250I/I263W | | Descriptor: | ZINC ION, organophosphorus hydrolase | | Authors: | Luo, X.J, Kong, X.D, Zhao, J, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Switching a newly discovered lactonase into an efficient and thermostable phosphotriesterase by simple double mutations His250Ile/Ile263Trp
Biotechnol.Bioeng., 111, 2014
|
|
6XBU
 
 | | polymerase domain of polymerase-theta | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*TP*CP*AP*TP*TP*G)-3'), DNA polymerase theta, ... | | Authors: | Chen, X, Pomerantz, R, Zhao, J. | | Deposit date: | 2020-06-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Pol theta reverse transcribes RNA and promotes RNA-templated DNA repair.
Sci Adv, 7, 2021
|
|
6WK7
 
 | | Crystal Structure Analysis of a poly(thymine) DNA duplex | | Descriptor: | 1,3,5-triazine-2,4,6-triamine, DNA (5'-D(*TP*TP*TP*TP*TP*T)-3') | | Authors: | Li, Q, Zhao, J, Liu, L, Mandal, S, Rizzuto, F.J, He, H, Wei, S, Jonchhe, S, Sleiman, H.F, Mao, H, Mao, C. | | Deposit date: | 2020-04-15 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | A poly(thymine)-melamine duplex for the assembly of DNA nanomaterials.
Nat Mater, 19, 2020
|
|
