5NSR
 
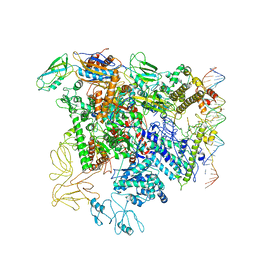 | | Cryo-EM structure of RNA polymerase-sigma54 holo enzyme with promoter DNA closed complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Darbari, V.C, Zhang, N, Buck, M, Zhang, X.D. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of RNA Polymerase Closed and Intermediate Complexes Reveal Mechanisms of DNA Opening and Transcription Initiation.
Mol. Cell, 67, 2017
|
|
5NSS
 
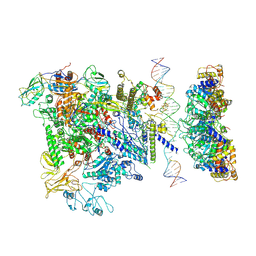 | | Cryo-EM structure of RNA polymerase-sigma54 holoenzyme with promoter DNA and transcription activator PspF intermedate complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Darbari, V.C, Zhang, N, Buck, M, Zhang, X.D. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of RNA Polymerase Closed and Intermediate Complexes Reveal Mechanisms of DNA Opening and Transcription Initiation.
Mol. Cell, 67, 2017
|
|
6R9B
 
 | |
6R9G
 
 | |
1JRU
 
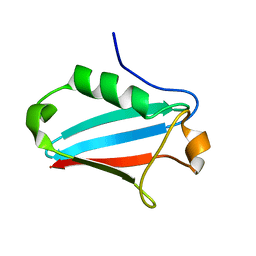 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 (ENERGY MINIMISED AVERAGE) | | Descriptor: | p47 protein | | Authors: | Yuan, X.M, Shaw, A, Zhang, X.D, Kondo, H, Lally, J, Freemont, P.S, Matthews, S.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
7QXI
 
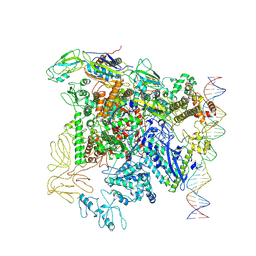 | |
7QWP
 
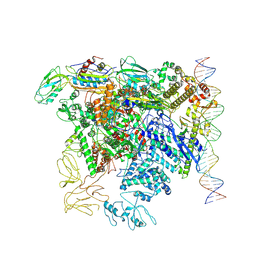 | | CryoEM structure of bacterial transcription close complex (RPc) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Ye, F.Z, Zhang, X.D. | | Deposit date: | 2022-01-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanisms of DNA opening revealed in AAA+ transcription complex structures.
Sci Adv, 8, 2022
|
|
7QV9
 
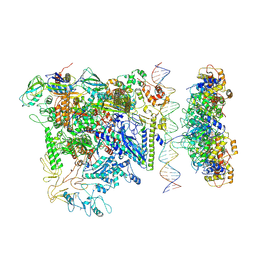 | |
4GKG
 
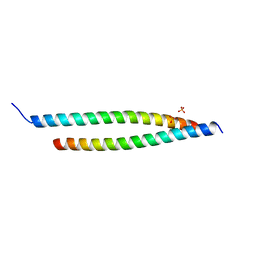 | | Crystal structure of the S-Helix Linker | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, PHOSPHATE ION | | Authors: | Liu, J.W, Lu, D, Sun, Y.J, Wen, J, Yang, Y, Yang, J.G, Wei, X.L, Zhang, X.D, Wang, Y.P. | | Deposit date: | 2012-08-11 | | Release date: | 2013-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of the S-Helix Linker
To be Published
|
|
6GH5
 
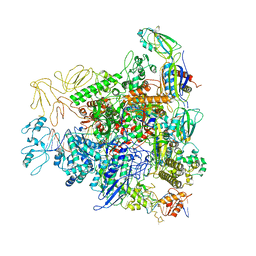 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme transcription open complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2018-05-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
6GFW
 
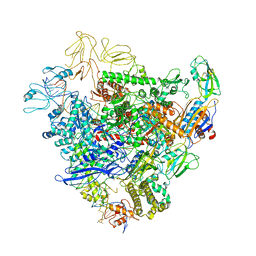 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme initial transcribing complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
6GH6
 
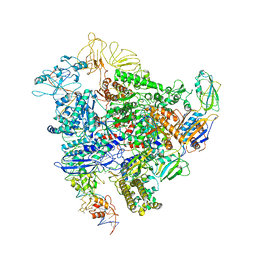 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme intermediate partially loaded complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2018-05-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
7BEG
 
 | | Structures of class I bacterial transcription complexes | | Descriptor: | Class I pacrA promoter non-template DNA, Class I pacrA promoter template DNA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Ye, F.Z, Hao, M, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
7BEF
 
 | | Structures of class II bacterial transcription complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Hao, M, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
