3ECR
 
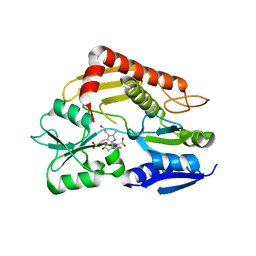 | | Structure of human porphobilinogen deaminase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Song, G, Li, Y, Cheng, C, Zhao, Y, Gao, A, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural insight into acute intermittent porphyria.
Faseb J., 23, 2009
|
|
1L8R
 
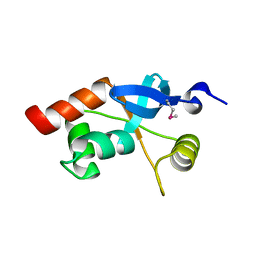 | | Structure of the Retinal Determination Protein Dachshund Reveals a DNA-Binding Motif | | Descriptor: | Dachshund | | Authors: | Kim, S.S, Zhang, R, Braunstein, S.E, Joachimiak, A, Cvekl, A, Hegde, R.S. | | Deposit date: | 2002-03-21 | | Release date: | 2002-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the retinal determination protein Dachshund reveals a DNA binding motif.
Structure, 10, 2002
|
|
3ZOA
 
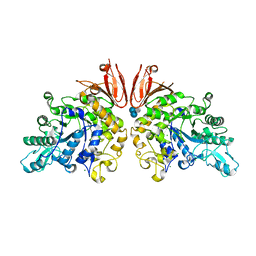 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
3ZO9
 
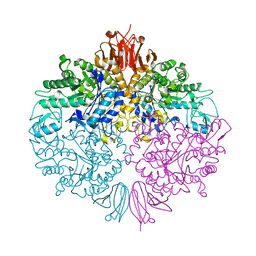 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
3GE2
 
 | | Crystal structure of putative lipoprotein SP_0198 from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein, ... | | Authors: | Kim, Y, Zhang, R, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Putative Lipoprotein SP_0198 from Streptococcus pneumoniae
To be Published
|
|
1YY6
 
 | | The Crystal Structure of the N-terminal domain of HAUSP/USP7 complexed with an EBNA1 peptide | | Descriptor: | Epstein-Barr nuclear antigen-1, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Saridakis, V, Sheng, Y, Sarkari, F, Holowaty, M, Shire, K, Nguyen, T, Zhang, R, Liao, J, Lee, W, Edwards, A.M, Arrowsmith, C.H, Frappier, L. | | Deposit date: | 2005-02-23 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.
Mol.Cell, 18, 2005
|
|
3HXT
 
 | | Structure of human MTHFS | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, NICKEL (II) ION | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3IJ8
 
 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | (2R,3S,4R,5R,6R)-2,6-difluoro-2-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol, 5-fluoro-alpha-L-idopyranose, CALCIUM ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
3IJ7
 
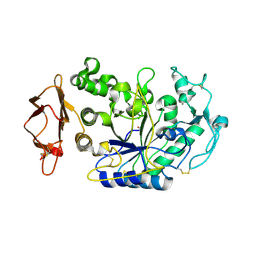 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | 4-O-methyl-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranosyl fluoride, 4-O-methyl-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-2)-5-fluoro-alpha-L-idopyranose, CALCIUM ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-03 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
3IJ9
 
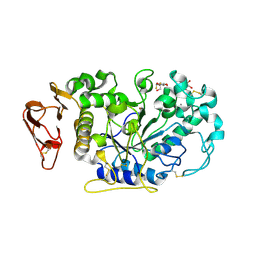 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | (2R,3S,4R,5R,6R)-2,6-difluoro-2-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
3J6E
 
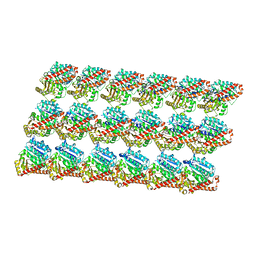 | | Energy minimized average structure of Microtubules stabilized by GmpCpp | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Alushin, G.M, Lander, G.C, Kellogg, E.H, Zhang, R, Baker, D, Nogales, E. | | Deposit date: | 2014-02-18 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | High-Resolution Microtubule Structures Reveal the Structural Transitions in alpha beta-Tubulin upon GTP Hydrolysis.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J6F
 
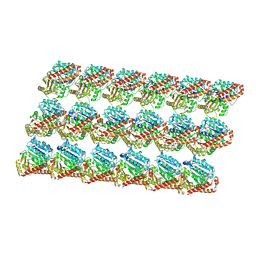 | | Minimized average structure of GDP-bound dynamic microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Alushin, G.M, Lander, G.C, Kellogg, E.H, Zhang, R, Baker, D, Nogales, E. | | Deposit date: | 2014-02-19 | | Release date: | 2014-06-04 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | High-Resolution Microtubule Structures Reveal the Structural Transitions in alpha beta-Tubulin upon GTP Hydrolysis.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J6G
 
 | | Minimized average structure of microtubules stabilized by taxol | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Alushin, G.M, Lander, G.C, Kellogg, E.H, Zhang, R, Baker, D, Nogales, E. | | Deposit date: | 2014-02-19 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | High-Resolution Microtubule Structures Reveal the Structural Transitions in alpha beta-Tubulin upon GTP Hydrolysis.
Cell(Cambridge,Mass.), 157, 2014
|
|
5W3F
 
 | | Yeast tubulin polymerized with GTP in vitro | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Howes, S.C, Geyer, E.A, LaFrance, B, Zhang, R, Kellogg, E.H, Westermann, S, Rice, L.M, Nogales, E. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural differences between yeast and mammalian microtubules revealed by cryo-EM.
J. Cell Biol., 216, 2017
|
|
5W3H
 
 | | Yeast microtubule stabilized with epothilone | | Descriptor: | EPOTHILONE A, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Howes, S.C, Geyer, E.A, LaFrance, B, Zhang, R, Kellogg, E.H, Westermann, S, Rice, L.M, Nogales, E. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural differences between yeast and mammalian microtubules revealed by cryo-EM.
J. Cell Biol., 216, 2017
|
|
5W3J
 
 | | Yeast microtubule stabilized with Taxol assembled from mutated tubulin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Howes, S.C, Geyer, E.A, LaFrance, B, Zhang, R, Kellogg, E.H, Westermann, S, Rice, L.M, Nogales, E. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural differences between yeast and mammalian microtubules revealed by cryo-EM.
J. Cell Biol., 216, 2017
|
|
5WTL
 
 | | Crystal structure of the periplasmic portion of outer membrane protein A (OmpA) from Capnocytophaga gingivalis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, OmpA family protein, ... | | Authors: | Dai, S, Tan, K, Ye, S, Zhang, R. | | Deposit date: | 2016-12-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability.
Cell Calcium, 66, 2017
|
|
5WTP
 
 | | Crystal structure of the C-terminal domain of outer membrane protein A (OmpA) from Capnocytophaga gingivalis | | Descriptor: | OmpA family protein, SULFATE ION | | Authors: | Dai, S, Tan, K, Ye, S, Zhang, R. | | Deposit date: | 2016-12-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability.
Cell Calcium, 66, 2017
|
|
5XJS
 
 | |
5XJT
 
 | |
5XJR
 
 | |
5XJQ
 
 | |
5XJU
 
 | |
5YHU
 
 | |
5ZHU
 
 | |
