5DUP
 
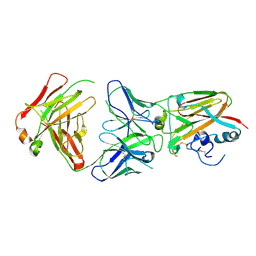 | | Influenza A virus H5 hemagglutinin globular head in complex with antibody AVFluIgG03 | | Descriptor: | AVFluIgG03 Heavy Chain, AVFluIgG03 Light Chain, Hemagglutinin | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUM
 
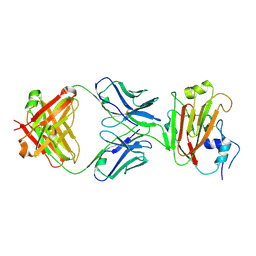 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody 65C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 65C6 Heavy Chain, 65C6 Light Chain, ... | | Authors: | Sun, J, Zuo, T, Wang, G, Zhou, P, Zhang, L, Wang, X. | | Deposit date: | 2015-09-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUR
 
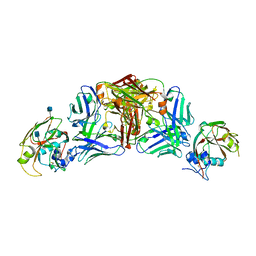 | | Influenza A virus H5 hemagglutinin globular head in complex with antibody 100F4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Antibody 100F4, Hemagglutinin, ... | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUT
 
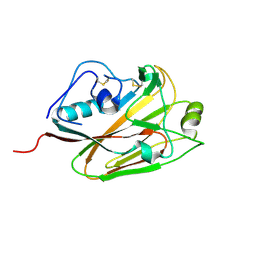 | | Influenza A virus H5 hemagglutinin globular head | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
7C0G
 
 | |
5YY5
 
 | | Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Zhang, S, Wang, P, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2017-12-08 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Definition of a Unique Neutralization Epitope on the Receptor-Binding Domain of MERS-CoV Spike Glycoprotein
Cell Rep, 24, 2018
|
|
6AHF
 
 | | CryoEM Reconstruction of Hsp104 N728A Hexamer | | Descriptor: | Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Zhang, X, Zhang, L, Zhang, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.78 Å) | | Cite: | Heat shock protein 104 (HSP104) chaperones soluble Tau via a mechanism distinct from its disaggregase activity.
J. Biol. Chem., 294, 2019
|
|
5YVF
 
 | | Crystal structure of BFA1 | | Descriptor: | BFA1 | | Authors: | Pu, H, Zhang, L, Duan, Z.K, Peng, L.W, Liu, L. | | Deposit date: | 2017-11-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Nucleus-Encoded Protein BFA1 Promotes Efficient Assembly of the Chloroplast ATP Synthase Coupling Factor 1.
Plant Cell, 30, 2018
|
|
5ZMD
 
 | | Crystal structure of FTO in complex with m6dA modified ssDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, DNA (5'-D(P*TP*CP*TP*(6MA)P*TP*AP*TP*CP*G)-3'), MANGANESE (II) ION, ... | | Authors: | Zhang, X, Wei, L.H, Luo, J, Xiao, Y, Liu, J, Zhang, W, Zhang, L, Jia, G.F. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5J19
 
 | | phospho-Pon binding-induced Plk1 dimerization | | Descriptor: | GLYCEROL, Phosphorylated peptide from Partner of Numb, Serine/threonine-protein kinase PLK1 | | Authors: | Zhu, K, Shan, Z, Zhang, L, Wen, W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phospho-Pon Binding-Mediated Fine-Tuning of Plk1 Activity
Structure, 24, 2016
|
|
5KC2
 
 | | Negative stain structure of Vps15/Vps34 complex | | Descriptor: | Phosphatidylinositol 3-kinase VPS34, Serine/threonine-protein kinase VPS15 | | Authors: | Kirsten, M.L, Zhang, L, Ohashi, Y, Perisic, O, Williams, R.L, Sachse, C. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
6CSV
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 63 kDa,Centrosomal protein of 152 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
6CSU
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 152 kDa, Centrosomal protein of 63 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
5DP4
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 3 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2S)-2-methyl-3-phenylpropanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5ZAT
 
 | | Crystal structure of 5-carboxylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(CAC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
5DP9
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 9 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(cyclobutylmethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP5
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 4 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2R,5S)-5-amino-2-(4-fluorobenzyl)-6-methyl-4-oxoheptanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2016-04-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP7
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 5 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-methylbutanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP6
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 7 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-cyclopropylpropanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DFZ
 
 | | Structure of Vps34 complex II from S. cerevisiae. | | Descriptor: | Nanobody binding S. cerevisiae Vps34, Phosphatidylinositol 3-kinase VPS34, Putative N-terminal domain of S. cerevisiae Vps30, ... | | Authors: | Rostislavleva, K, Soler, N, Ohashi, Y, Zhang, L, Williams, R.L. | | Deposit date: | 2015-08-27 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes.
Science, 350, 2015
|
|
5DP3
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 2 | | Descriptor: | 3C proteinase, ethyl (4S)-5-[(3S)-2-oxopyrrolidin-3-yl]-4-[(3-phenylpropanoyl)amino]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP8
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 8 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(2-cyclopropylethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DPA
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 6 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-acetyl-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5ZAS
 
 | | Crystal structure of 5-formylcytosine containing decamer dsDNA | | Descriptor: | BICARBONATE ION, DNA (5'-D(*CP*CP*AP*GP*(5FC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
