3B8V
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221K | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8U
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
6AHF
 
 | | CryoEM Reconstruction of Hsp104 N728A Hexamer | | Descriptor: | Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Zhang, X, Zhang, L, Zhang, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-02-13 | | Last modified: | 2019-04-10 | | Method: | ELECTRON MICROSCOPY (6.78 Å) | | Cite: | Heat shock protein 104 (HSP104) chaperones soluble Tau via a mechanism distinct from its disaggregase activity.
J. Biol. Chem., 294, 2019
|
|
8CWT
 
 | |
8CYF
 
 | | WhiB3 bound to SigmaAr4-RNAP Beta flap tip chimera and DNA | | Descriptor: | CACODYLATE ION, DNA (5'-D(*CP*AP*CP*CP*AP*CP*AP*AP*CP*CP*GP*AP*TP*TP*TP*T)-3'), DNA (5'-D(*GP*AP*AP*AP*AP*TP*CP*GP*GP*TP*TP*GP*TP*GP*GP*T)-3'), ... | | Authors: | Wan, T, Zhang, L.M. | | Deposit date: | 2022-05-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis of DNA binding by the WhiB-like transcription factor WhiB3 in Mycobacterium tuberculosis.
J.Biol.Chem., 299, 2023
|
|
6ONO
 
 | | Complex structure of WhiB1 and region 4 of SigA in C2221 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
3EAX
 
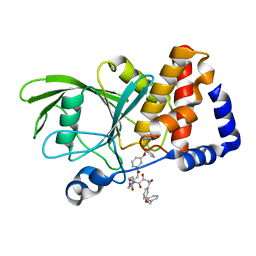 | | Crystal structure PTP1B complex with small molecule compound LZP-6 | | Descriptor: | 4,4'-piperazine-1,4-diylbis{1-[3-(benzyloxy)phenyl]-4-oxobutane-1,3-dione}, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Zhang, Z.-Y, Liu, S, Zhang, L.-F, Yu, X, Xue, T, Gunawan, A.M, Long, Y.-Q. | | Deposit date: | 2008-08-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting inactive enzyme conformation: aryl diketoacid derivatives as a new class of PTP1B inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
8JOO
 
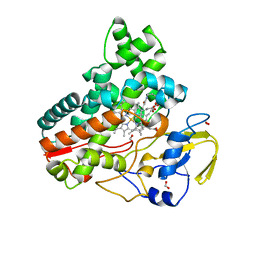 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate ikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-08 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JNC
 
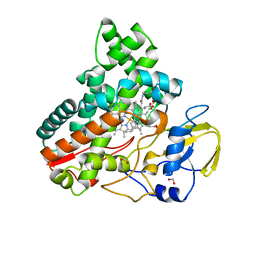 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-maltophilin | | Descriptor: | (1Z,3E,5S,8R,9S,10S,11R,13R,15R,16S,18Z,24S,25S)-11-ethyl-2,24-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-7,20,27,28-tetraone, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JUA
 
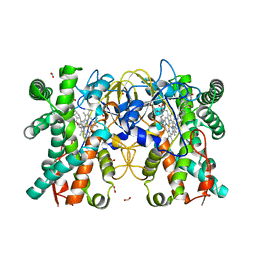 | | Multifunctional cytochrome P450 enzyme IkaD from Streptomyces sp. ZJ306, in complex with epoxyikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,13R,15S,16R,17S,19Z,26S)-11-ethyl-2-hydroxy-10-methyl-22,27-diaza-14 oxahexacyclo[24.2.1.05,17.07,16.013,15.08,12]nonacosa-1(2),3,19-triene-21,28,29-trione, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.00001121 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8SK5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing VHH 7A9 bound to the spike receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, anti-SARS-CoV-2 receptor binding domain VHH | | Authors: | Noland, C.L, Pande, K, Zhang, L, Zhou, H, Galli, J, Eddins, M, Gomez-Llorente, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Discovery and multimerization of cross-reactive single-domain antibodies against SARS-like viruses to enhance potency and address emerging SARS-CoV-2 variants.
Sci Rep, 13, 2023
|
|
6JEP
 
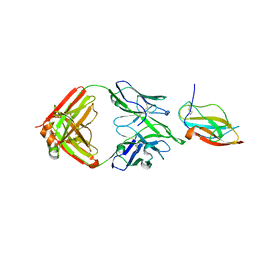 | | Structure of a neutralizing antibody bound to the Zika envelope protein domain III | | Descriptor: | Genome polyprotein, heavy chain of Fab ZK2B10, light chain of Fab ZK2B10 | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-07 | | Release date: | 2019-05-15 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
4EHR
 
 | | Crystal structure of Bcl-Xl complex with 4-(5-butyl-3-(hydroxymethyl)-1-phenyl-1h-pyrazol-4-yl)-3-(3,4-dihydro-2(1h)-isoquinolinylcarbonyl)-n-((2-(trimethylsilyl)ethyl)sulfonyl)benzamide | | Descriptor: | 4-[5-butyl-3-(hydroxymethyl)-1-phenyl-1H-pyrazol-4-yl]-3-(3,4-dihydroisoquinolin-2(1H)-ylcarbonyl)-N-{[2-(trimethylsilyl)ethyl]sulfonyl}benzamide, Bcl-2-like protein 1, IMIDAZOLE | | Authors: | Schroeder, G.M, Wei, D, Banfi, P, Cai, Z, Lippy, J, Menichincheri, M, Modugno, M, Naglich, J, Penhallow, B, Perez, H.L, Sack, J, Schmidt, R.J, Tebben, A, Yan, C, Zhang, L, Galvani, A, Lombardo, L.J, Borzilleri, R.M. | | Deposit date: | 2012-04-03 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyrazole and pyrimidine phenylacylsulfonamides as dual Bcl-2/Bcl-xL antagonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6JFH
 
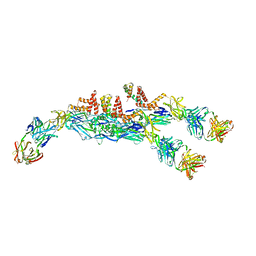 | | The asymmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | Descriptor: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural E protein, ... | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6E8J
 
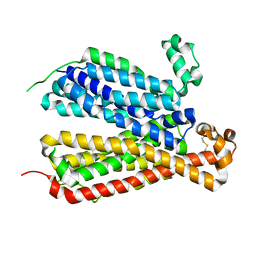 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Occupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6E9C
 
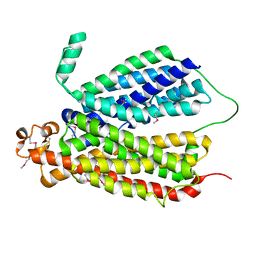 | | Selenomethionine Derivative Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
7QBA
 
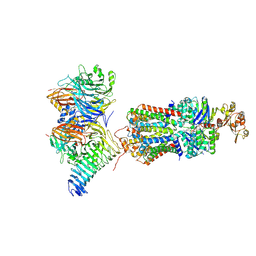 | | CryoEM structure of the ABC transporter NosDFY complexed with nitrous oxide reductase NosZ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zipfel, S, Mueller, C, Topitsch, A, Lutz, M, Zhang, L, Einsle, O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-03 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
5XPV
 
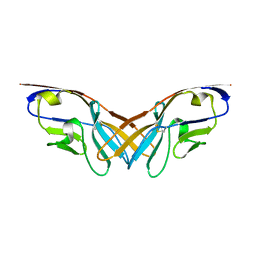 | | Structure of the V domain of amphioxus IgVJ-C2 | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
5XPW
 
 | | Structure of amphioxus IgVJ-C2 molecule | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
8JNO
 
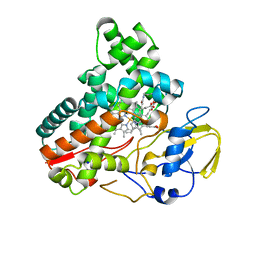 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-deOH-HSAF | | Descriptor: | (1Z,3E,5S,7S,8R,9S,10S,11R,13R,15R,16S,18Z,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7REF
 
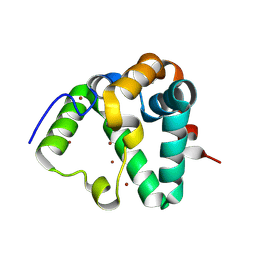 | | Structure of MS3494 from Mycobacterium smegmatis | | Descriptor: | BROMIDE ION, MS3494 | | Authors: | Kent, J.E, Aleshin, A.E, Zhang, L, Niederweis, M, Marassi, F.M. | | Deposit date: | 2021-07-12 | | Release date: | 2021-08-18 | | Last modified: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A periplasmic cinched protein is required for siderophore secretion and virulence of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
5Z6P
 
 | | The crystal structure of an agarase, AgWH50C | | Descriptor: | B-agarase | | Authors: | Mao, X, Zhou, J, Zhang, P, Zhang, L, Zhang, J, Li, Y. | | Deposit date: | 2018-01-24 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Structure-based design of agarase AgWH50C from Agarivorans gilvus WH0801 to enhance thermostability.
Appl. Microbiol. Biotechnol., 103, 2019
|
|
1L0L
 
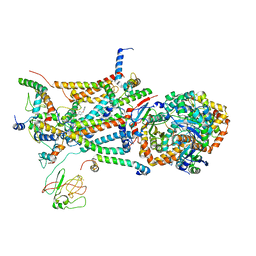 | | structure of bovine mitochondrial cytochrome bc1 complex with a bound fungicide famoxadone | | Descriptor: | Cytochrome B, Cytochrome c1, heme protein, ... | | Authors: | Gao, X, Wen, X, Yu, C.A, Esser, L, Tsao, S, Quinn, B, Zhang, L, Yu, L, Xia, D. | | Deposit date: | 2002-02-11 | | Release date: | 2003-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of Mitochondrial Cytochrome bc1 in Complex with Famoxadone: The Role of Aromatic-Aromatic Interaction in Inhibition
Biochemistry, 41, 2003
|
|
1L0N
 
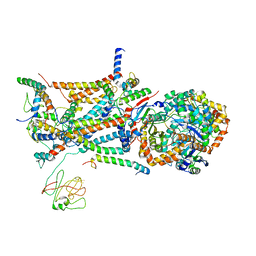 | | native structure of bovine mitochondrial cytochrome bc1 complex | | Descriptor: | Cytochrome B, Cytochrome c1, heme protein, ... | | Authors: | Gao, X, Wen, X, Yu, C.A, Esser, L, Tsao, S, Quinn, B, Zhang, L, Yu, L, Xia, D. | | Deposit date: | 2002-02-11 | | Release date: | 2003-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of Mitochondrial Cytochrome bc1 in Complex with Famoxadone: The Role of Aromatic-Aromatic Interaction in Inhibition
Biochemistry, 41, 2003
|
|
6JFI
 
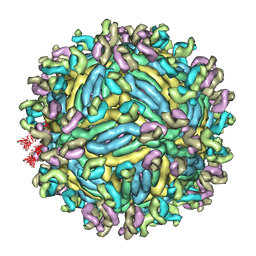 | | The symmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | Descriptor: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural protein E, ... | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
