3J17
 
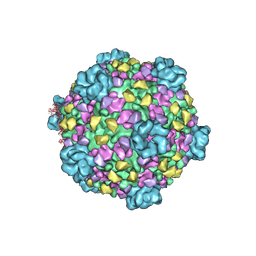 | | Structure of a transcribing cypovirus by cryo-electron microscopy | | Descriptor: | Structural protein VP3, Structural protein VP5, VP1 | | Authors: | Yang, C, Ji, G, Liu, H, Zhang, K, Liu, G, Sun, F, Zhu, P, Cheng, L. | | Deposit date: | 2011-12-25 | | Release date: | 2012-04-04 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a transcribing cypovirus.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3K6J
 
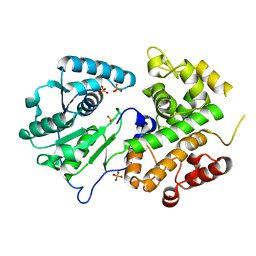 | | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans | | Descriptor: | PHOSPHATE ION, Protein F01G10.3, confirmed by transcript evidence, ... | | Authors: | Ouyang, Z, Zhang, K, Zhai, Y, Lu, J, Sun, F. | | Deposit date: | 2009-10-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans
To be Published
|
|
5AFU
 
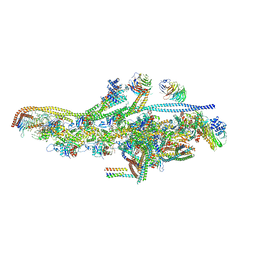 | | Cryo-EM structure of dynein tail-dynactin-BICD2N complex | | Descriptor: | ACTIN, CYTOPLASMIC 1, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Urnavicius, L, Zhang, K, Diamant, A.G, Motz, C, Schlager, M.A, Yu, M, Patel, N.A, Robinson, C.V, Carter, A.P. | | Deposit date: | 2015-01-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The Structure of the Dynactin Complex and its Interaction with Dynein.
Science, 347, 2015
|
|
4HND
 
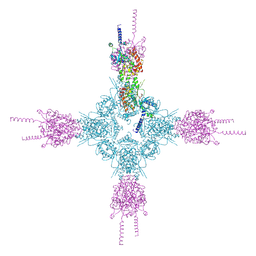 | | Crystal structure of the catalytic domain of Selenomethionine substituted human PI4KIIalpha in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | Deposit date: | 2012-10-19 | | Release date: | 2014-04-09 | | Last modified: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
4HNE
 
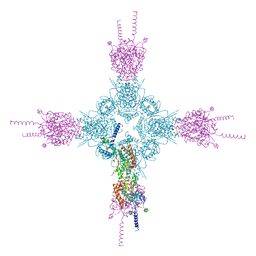 | | Crystal structure of the catalytic domain of human type II alpha Phosphatidylinositol 4-kinase (PI4KIIalpha) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | Deposit date: | 2012-10-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
6BCG
 
 | | I-LtrI A28G bound to cognate substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (26-MER), Ribosomal protein 3/homing endonuclease-like fusion protein | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
6BD0
 
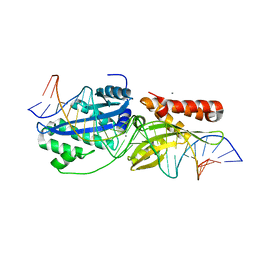 | | I-OnuI K227Y, D236A bound to cognate substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (25-MER), Ribosomal protein 3/homing endonuclease-like protein fusion | | Authors: | Brown, C, Zhang, K, Laforet, M, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | I-OnuI K227Y, D236A bound to cognate substrate (pre-cleavage complex)
To Be Published
|
|
6BDB
 
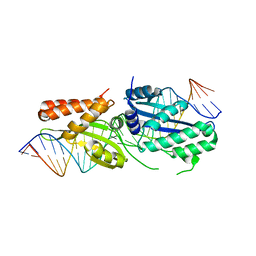 | | I-OnuI K227Y, D236A bound to A3G substrate (pre-cleavage complex) | | Descriptor: | DNA (26-MER), Ribosomal protein 3/homing endonuclease-like protein fusion | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Laforet, M, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | I-OnuI K227Y, D236A bound to A3G substrate (pre-cleavage complex)
To Be Published
|
|
6BCF
 
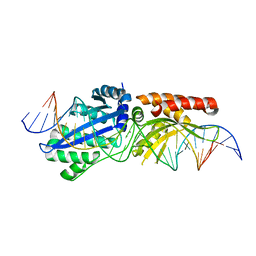 | | I-LtrI G183A bound to cognate substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (26-MER), Ribosomal protein 3/homing endonuclease-like fusion protein | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
6BCH
 
 | | I-LtrI E29D bound to cognate substrate (nicked complex) | | Descriptor: | DNA (26-MER), DNA (5'-D(*GP*GP*TP*CP*TP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*GP*GP*AP*GP*CP*AP*TP*TP*T)-3'), ... | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | I-LtrI E29D bound to cognate substrate (nicked complex)
To Be Published
|
|
6BCT
 
 | | I-LtrI E184D bound to non-cognate C4 substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (26-MER), DNA (27-MER), ... | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
6BDA
 
 | | Wild-type I-OnuI bound to A3G substrate (post-cleavage complex) | | Descriptor: | Cleaved Cognate DNA strand, +11 sense, Cleaved cognate DNA strand, ... | | Authors: | Brown, C, Zhang, K, Laforet, M, McMurrough, T, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Wild-type I-OnuI bound to A3G substrate (post-cleavage complex)
To Be Published
|
|
6BCE
 
 | | Wild-type I-LtrI bound to cognate substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (27-MER), Ribosomal protein 3/homing endonuclease-like fusion protein | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
6BDG
 
 | | HFQ monomer in spacegroup p6 at 1.93 angstrom resolution | | Descriptor: | RNA-binding protein Hfq | | Authors: | Brown, C, Zhang, K, Seo, C, Ellis, M.J, Hanniford, D.B, Junop, M. | | Deposit date: | 2017-10-23 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | HFQ monomer in spacegroup p6 at 1.93 angstrom resolution
To Be Published
|
|
6BCI
 
 | | Wild-type I-LtrI bound to non-cognate C4 substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (27-MER), Ribosomal protein 3/homing endonuclease-like fusion protein | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
4EGT
 
 | | Crystal structure of major capsid protein P domain from rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Wang, X, Xu, F, Zhang, K, Zhai, Y, Sun, F. | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
4CKH
 
 | | Helical reconstruction of ACAP1(BAR-PH domain) decorated membrane tubules by cryo-electron microscopy | | Descriptor: | ARF-GAP WITH COILED-COIL, ANK REPEAT AND PH DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Pang, X.Y, Fan, J, Zhang, Y, Zhang, K, Gao, B.Q, Ma, J, Li, J, Deng, Y.C, Zhou, Q.J, Hsu, V, Sun, F. | | Deposit date: | 2014-01-06 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | A Ph Domain in Acap1 Possesses Key Features of the Bar Domain in Promoting Membrane Curvature.
Dev.Cell, 31, 2014
|
|
4CKG
 
 | | Helical reconstruction of ACAP1(BAR-PH domain) decorated membrane tubules by cryo-electron microscopy | | Descriptor: | ARF-GAP WITH COILED-COIL, ANK REPEAT AND PH DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Pang, X.Y, Fan, J, Zhang, Y, Zhang, K, Gao, B.Q, Ma, J, Li, J, Deng, Y.C, Zhou, Q.J, Hsu, V, Sun, F. | | Deposit date: | 2014-01-06 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | A Ph Domain in Acap1 Possesses Key Features of the Bar Domain in Promoting Membrane Curvature.
Dev.Cell, 31, 2014
|
|
4EJR
 
 | | Crystal structure of major capsid protein S domain from rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Xu, F, Ma, J, Zhang, K, Wang, X, Sun, F. | | Deposit date: | 2012-04-07 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
4MBQ
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Robinson, H, Wolfram, F, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
7ARM
 
 | | LolCDE in complex with lipoprotein and LolA | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARL
 
 | | LolCDE in complex with lipoprotein and ADP | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, ADENOSINE-5'-DIPHOSPHATE, LPP, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARJ
 
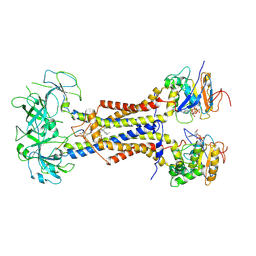 | | LolCDE in complex with lipoprotein and AMPPNP complex undimerized form | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARI
 
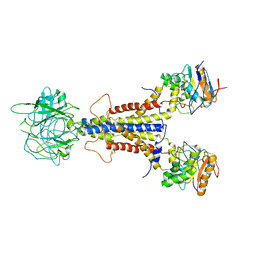 | | LolCDE apo structure | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARK
 
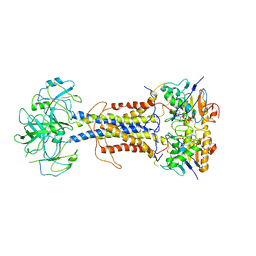 | | LolCDE in complex with AMP-PNP in the closed NBD state | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
