8JAL
 
 | | Structure of CRL2APPBP2 bound with RxxGP degron (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAQ
 
 | | Structure of CRL2APPBP2 bound with RxxGP degron (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAS
 
 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAU
 
 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAV
 
 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5EWP
 
 | | ARO (armadillo repeats only protein) from Plasmodium falciparum | | Descriptor: | ARO (armadillo repeats only protein) | | Authors: | Brown, C, Zhang, K, Emery, J, Prusty, D, Wetzel, J, Heincke, D, Gilberger, T, Junop, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ARO (armadillo repeats only protein) from Plasmodium falciparum
To Be Published
|
|
3F7O
 
 | | Crystal structure of Cuticle-Degrading Protease from Paecilomyces lilacinus (PL646) | | Descriptor: | (MSU)(ALA)(ALA)(PRO)(VAL), CALCIUM ION, Serine protease | | Authors: | Liang, L, Lou, Z, Meng, Z, Rao, Z, Zhang, K. | | Deposit date: | 2008-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
3F7M
 
 | | Crystal structure of apo Cuticle-Degrading Protease (ver112) from Verticillium psalliotae | | Descriptor: | Alkaline serine protease ver112 | | Authors: | Liang, L, Lou, Z, Ye, F, Meng, Z, Rao, Z, Zhang, K. | | Deposit date: | 2008-11-09 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
8J60
 
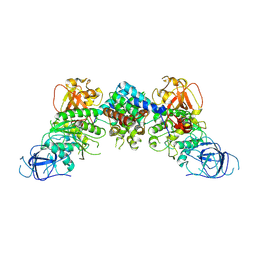 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 protein, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
8J5Y
 
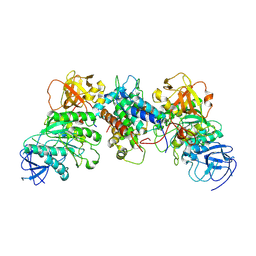 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 isoform 1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
8JVB
 
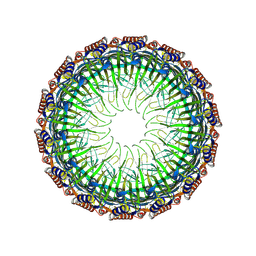 | | Cryo-EM structure of the Type II secretion system protein from Acidithiobacillus caldus | | Descriptor: | Type II and III secretion system protein | | Authors: | Liu, R.H, Zhang, K, Feng, Q.S, Dai, X, Fu, Y, Li, Y. | | Deposit date: | 2023-06-28 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Cryo-EM structure of the Type II secretion system protein from Acidithiobacillus caldus
To Be Published
|
|
4Y9L
 
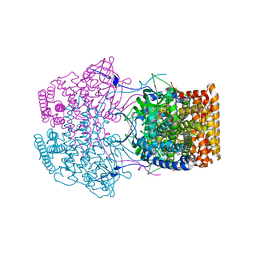 | | Crystal Structure of Caenorhabditis elegans ACDH-11 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Protein ACDH-11, isoform b | | Authors: | Li, Z.J, Zhai, Y.J, Zhang, K, Sun, F. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Acyl-CoA Dehydrogenase Drives Heat Adaptation by Sequestering Fatty Acids
Cell, 161, 2015
|
|
6K61
 
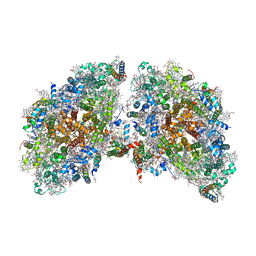 | | Cryo-EM structure of the tetrameric photosystem I from a heterocyst-forming cyanobacterium Anabaena sp. PCC7120 | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Zheng, L, Li, Y, Li, X, Zhong, Q, Li, N, Zhang, K, Zhang, Y, Chu, H, Ma, C, Li, G, Zhao, J, Gao, N. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural and functional insights into the tetrameric photosystem I from heterocyst-forming cyanobacteria.
Nat.Plants, 5, 2019
|
|
3T9K
 
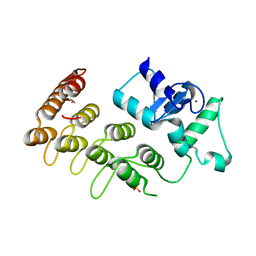 | | Crystal Structure of ACAP1 C-portion mutant S554D fused with integrin beta1 peptide | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1,Peptide from Integrin beta-1, SULFATE ION, ... | | Authors: | Sun, F, Pang, X, Zhang, K, Ma, J, Zhou, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
8D0A
 
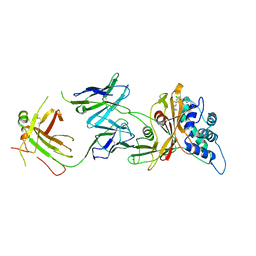 | | Crystal structure of human USP30 in complex with a covalent inhibitor 829 and a Fab | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 30, ZINC ION, mouse anti-huUSP30 Fab heavy chain, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | TBD
To Be Published
|
|
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | Descriptor: | RNA (387-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3U9G
 
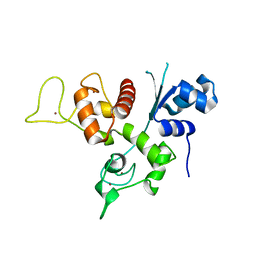 | | Crystal structure of the Zinc finger antiviral protein | | Descriptor: | ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Chen, S, Xu, Y, Zhang, K, Wang, X, Sun, J, Gao, G, Liu, Y. | | Deposit date: | 2011-10-18 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7XML
 
 | | Cryo-EM structure of PEIP-Bs_enolase complex | | Descriptor: | Enolase, MAGNESIUM ION, Putative gene 60 protein | | Authors: | Li, S, Zhang, K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage protein PEIP is a potent Bacillus subtilis enolase inhibitor.
Cell Rep, 40, 2022
|
|
8J72
 
 | | Crystal structure of mammalian Trim71 in complex with lncRNA Trincr1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM71, lncRNA Trincr1 | | Authors: | Shi, F.D, Zhang, K, Che, S.Y, Zhi, S.X, Yang, N. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Molecular mechanism governing RNA-binding property of mammalian TRIM71 protein.
Sci Bull (Beijing), 69, 2024
|
|
7BZ0
 
 | | complex structure of alginate lyase AlyF-OU02 with G6 | | Descriptor: | Alginate lyase AlyF-OU02, CALCIUM ION, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Liu, W, Lyu, Q, Zhang, K. | | Deposit date: | 2020-04-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the substrate-binding cleft of AlyF reveal the first long-chain alginate-binding mode.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8K3Y
 
 | | The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Zhang, K, Chang, C.I. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7C49
 
 | | nicA2 with cofactor FAD and substrate nicotine | | Descriptor: | 5-[(2S)-1-methylpyrrolidin-2-yl]pyridin-2-ol, Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xu, P, Zhang, K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Deceleration Regulates Toxicant Release to Prevent Cell Damage in Pseudomonas putida S16 (DSM 28022).
Mbio, 11, 2020
|
|
