7XN1
 
 | | Crystal structure of human acetylcholinesterase in complex with tacrine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Acetylcholinesterase, ... | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2022-04-27 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
5HSJ
 
 | | Structure of tyrosine decarboxylase complex with PLP at 1.9 Angstroms resolution | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase | | Authors: | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | Deposit date: | 2016-01-25 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
3K6J
 
 | | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans | | Descriptor: | PHOSPHATE ION, Protein F01G10.3, confirmed by transcript evidence, ... | | Authors: | Ouyang, Z, Zhang, K, Zhai, Y, Lu, J, Sun, F. | | Deposit date: | 2009-10-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans
To be Published
|
|
3J17
 
 | | Structure of a transcribing cypovirus by cryo-electron microscopy | | Descriptor: | Structural protein VP3, Structural protein VP5, VP1 | | Authors: | Yang, C, Ji, G, Liu, H, Zhang, K, Liu, G, Sun, F, Zhu, P, Cheng, L. | | Deposit date: | 2011-12-25 | | Release date: | 2012-04-04 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a transcribing cypovirus.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3JUE
 
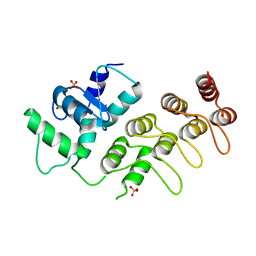 | | Crystal Structure of ArfGAP and ANK repeat domain of ACAP1 | | Descriptor: | ARFGAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | Authors: | Pang, X, Zhang, K, Ma, J, Zhou, Q, Sun, F. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
3G6L
 
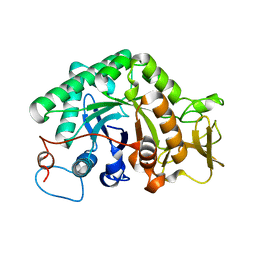 | |
3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | Descriptor: | CAFFEINE, Chitinase | | Authors: | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | Deposit date: | 2009-02-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | Descriptor: | RNA (387-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6BD0
 
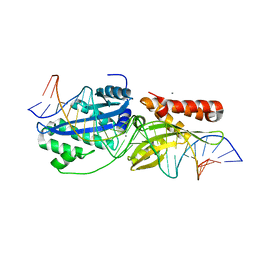 | | I-OnuI K227Y, D236A bound to cognate substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (25-MER), Ribosomal protein 3/homing endonuclease-like protein fusion | | Authors: | Brown, C, Zhang, K, Laforet, M, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | I-OnuI K227Y, D236A bound to cognate substrate (pre-cleavage complex)
To Be Published
|
|
6BDB
 
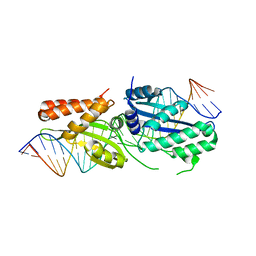 | | I-OnuI K227Y, D236A bound to A3G substrate (pre-cleavage complex) | | Descriptor: | DNA (26-MER), Ribosomal protein 3/homing endonuclease-like protein fusion | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Laforet, M, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | I-OnuI K227Y, D236A bound to A3G substrate (pre-cleavage complex)
To Be Published
|
|
6BCT
 
 | | I-LtrI E184D bound to non-cognate C4 substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (26-MER), DNA (27-MER), ... | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
6BCE
 
 | | Wild-type I-LtrI bound to cognate substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (27-MER), Ribosomal protein 3/homing endonuclease-like fusion protein | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
6BCI
 
 | | Wild-type I-LtrI bound to non-cognate C4 substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (27-MER), Ribosomal protein 3/homing endonuclease-like fusion protein | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
7XML
 
 | | Cryo-EM structure of PEIP-Bs_enolase complex | | Descriptor: | Enolase, MAGNESIUM ION, Putative gene 60 protein | | Authors: | Li, S, Zhang, K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage protein PEIP is a potent Bacillus subtilis enolase inhibitor.
Cell Rep, 40, 2022
|
|
7D3U
 
 | | Structure of Mrp complex from Dietzia sp. DQ12-45-1b | | Descriptor: | Cation antiporter, DODECYL-BETA-D-MALTOSIDE, Monovalent Na+/H+ antiporter subunit A, ... | | Authors: | Li, B, Zhang, K.D, Wu, X.L, Zhang, X.C. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7W16
 
 | | Complex structure of alginate lyase AlyV with M8 | | Descriptor: | GLYCEROL, alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W18
 
 | | Complex structure of alginate lyase PyAly with M5 | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W13
 
 | | Complex structure of alginate lyase PyAly with M8 | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
8J5Y
 
 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 isoform 1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
8J60
 
 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 protein, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
7E3D
 
 | | Crystal structure of human acetylcholinesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Acetylcholinesterase | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2021-02-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
7E3H
 
 | | Crystal structure of human acetylcholinesterase in complex with donepezil | | Descriptor: | 1-BENZYL-4-[(5,6-DIMETHOXY-1-INDANON-2-YL)METHYL]PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2021-02-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
