5V93
 
 | | Cryo-EM structure of the 70S ribosome from Mycobacterium tuberculosis bound with Capreomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Yang, K, Chang, J.-Y, Cui, Z, Li, X, Meng, R, Duan, L, Thongchol, J, Jakana, J, Huwe, C, Sacchettini, J, Zhang, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-20 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
2JZ3
 
 | | SOCS box elonginBC ternary complex | | Descriptor: | Suppressor of cytokine signaling 3, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2 | | Authors: | Babon, J.J, Sabo, J, Soetopo, A, Yao, S, Bailey, M.F, Zhang, J, Nicola, N.A, Norton, R.S. | | Deposit date: | 2007-12-27 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The SOCS box domain of SOCS3: structure and interaction with the elonginBC-cullin5 ubiquitin ligase
J.Mol.Biol., 381, 2008
|
|
3EMN
 
 | | The Crystal Structure of Mouse VDAC1 at 2.3 A resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Ujwal, R, Cascio, D, Colletier, J.-P, Faham, S, Zhang, J, Toro, L, Ping, P, Abramson, J. | | Deposit date: | 2008-09-24 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EB7
 
 | | Crystal Structure of Insecticidal Delta-Endotoxin Cry8Ea1 from Bacillus Thuringiensis at 2.2 Angstroms Resolution | | Descriptor: | ACETATE ION, Insecticidal Delta-Endotoxin Cry8Ea1, SULFATE ION | | Authors: | Guo, S, Ye, S, Song, F, Zhang, J, Wei, L, Shu, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Bacillus thuringiensis Cry8Ea1: An insecticidal toxin toxic to underground pests, the larvae of Holotrichia parallela.
J.Struct.Biol., 168, 2009
|
|
2M80
 
 | | Solution structure of yeast dithiol glutaredoxin Grx8 | | Descriptor: | Glutaredoxin-8 | | Authors: | Tang, Y, Zhang, J, Yu, J, Wu, J, Zhou, C.Z, Shi, Y. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-guided activity enhancement and catalytic mechanism of yeast grx8
Biochemistry, 53, 2014
|
|
5V7Q
 
 | | Cryo-EM structure of the large ribosomal subunit from Mycobacterium tuberculosis bound with a potent linezolid analog | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Yang, K, Chang, J.-Y, Cui, Z, Zhang, J. | | Deposit date: | 2017-03-20 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
8WJL
 
 | | Cryo-EM structure of 6-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.15 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8WJN
 
 | | Cryo-EM structure of 6-subunit Smc5/6 head region | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosomes element 1, Non-structural maintenance of chromosomes element 4, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.58 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8WJO
 
 | | Cryo-EM structure of 8-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2GIB
 
 | | Crystal structure of the SARS coronavirus nucleocapsid protein dimerization domain | | Descriptor: | Nucleocapsid protein, SULFATE ION | | Authors: | Yu, I.M, Oldham, M.L, Zhang, J, Chen, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae.
J.Biol.Chem., 281, 2006
|
|
2JXW
 
 | |
2H60
 
 | | Solution Structure of Human Brg1 Bromodomain | | Descriptor: | Probable global transcription activator SNF2L4 | | Authors: | Shen, W, Xu, C, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2006-05-30 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails
Biochemistry, 46, 2007
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
2PPH
 
 | | solution structure of human MEKK3 PB1 domain | | Descriptor: | Mitogen-activated protein kinase kinase kinase 3 | | Authors: | Hu, Q, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insight into the Binding Properties of MEKK3 PB1 to MEK5 PB1 from Its Solution Structure.
Biochemistry, 46, 2007
|
|
2LKZ
 
 | | Solution structure of the second RRM domain of RBM5 | | Descriptor: | RNA-binding protein 5 | | Authors: | Song, Z, Wu, P, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2011-10-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RRM domain of RBM5 and its unusual binding characters for different RNA targets
Biochemistry, 2012
|
|
5VM7
 
 | | Pseudo-atomic model of the MurA-A2 complex | | Descriptor: | Maturation protein A2, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7VTC
 
 | | Crystal structure of MERS main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53865623 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLQ
 
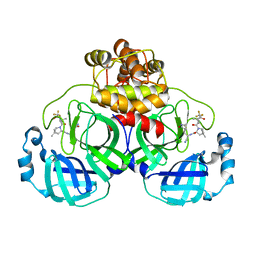 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P212121 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.939106 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLO
 
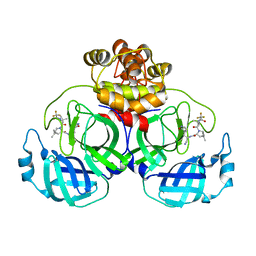 | | Crystal structure of SARS coronavirus main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.0227 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P1211 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.50251937 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
5VLZ
 
 | | Backbone model for phage Qbeta capsid | | Descriptor: | Capsid protein, Maturation protein A2 | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VLY
 
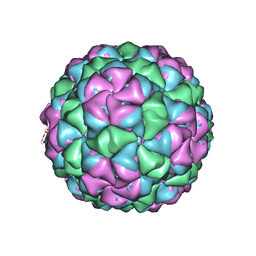 | | Asymmetric unit for the coat proteins of phage Qbeta | | Descriptor: | Capsid protein | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1C39
 
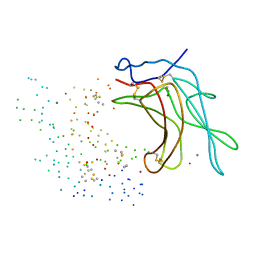 | | STRUCTURE OF CATION-DEPENDENT MANNOSE 6-PHOSPHATE RECEPTOR BOUND TO PENTAMANNOSYL PHOSPHATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose, CATION-DEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, ... | | Authors: | Olson, L.J, Zhang, J, Lee, Y.C, Dahms, N.M, Kim, J.J.-P. | | Deposit date: | 1999-07-25 | | Release date: | 2000-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for recognition of phosphorylated high mannose oligosaccharides by the cation-dependent mannose 6-phosphate receptor.
J.Biol.Chem., 274, 1999
|
|
2L43
 
 | | Structural basis for histone code recognition by BRPF2-PHD1 finger | | Descriptor: | Histone H3.3,LINKER,Bromodomain-containing protein 1, ZINC ION | | Authors: | Qin, S, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2010-10-01 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of unmodified histone H3 by the first PHD finger of Bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases MOZ and MORF
To be Published
|
|
2KXJ
 
 | | Solution structure of UBX domain of human UBXD2 protein | | Descriptor: | UBX domain-containing protein 4 | | Authors: | Wu, Q, Huang, H, Zhang, J, Hu, Q, Wu, J, Shi, Y. | | Deposit date: | 2010-05-06 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution strcture of UBX domain of human UBXD2 protein
To be Published
|
|
