7UPY
 
 | | An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPX
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SP1-77 Fab heavy chain, SP1-77 Fab light chain, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPW
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
5X16
 
 | | Sirt6 apo structure | | Descriptor: | GLYCEROL, NAD-dependent protein deacetylase sirtuin-6, TERTIARY-BUTYL ALCOHOL, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-01-24 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Sirt6 apo structure
To Be Published
|
|
5Z8H
 
 | | APC with an inhibitor | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, Peptide inhibitor | | Authors: | Zhang, J, Yang, X.Y, Song, K. | | Deposit date: | 2018-01-31 | | Release date: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | APC with an inhibitor
To Be Published
|
|
5ZY9
 
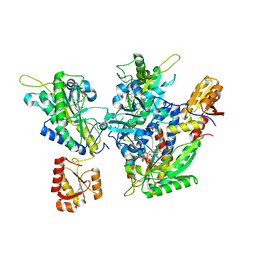 | | Structural basis for a tRNA synthetase | | Descriptor: | (1R,2R)-2-[(2S,4E,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadeca-4,6-dien-2-yl]cyclopentanecarboxylic acid, Threonyl-tRNA synthase, ZINC ION | | Authors: | Zhang, J. | | Deposit date: | 2018-05-23 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for inhibition of
translational functions on a tRNA synthetase
To Be Published
|
|
2NWM
 
 | |
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
8FDW
 
 | | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | Authors: | Zhang, J, Shi, W, Cai, Y.F, Zhu, H.S, Peng, H.Q, Voyer, J, Volloch, S.R, Cao, H, Mayer, M.L, Song, K.K, Xu, C, Lu, J.M, Chen, B. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane.
Nature, 619, 2023
|
|
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6WPI
 
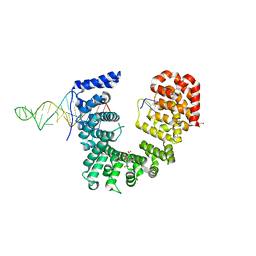 | | Crystal structure of Nop9 in complex with ITS1 RNA | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, GLYCEROL, Nucleolar protein 9, ... | | Authors: | Zhang, J, Qiu, C, Hall, T.M.T. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Nop9 recognizes structured and single-stranded RNA elements of preribosomal RNA.
Rna, 26, 2020
|
|
2FGF
 
 | |
8GSJ
 
 | | APC-Asef tripeptide inhibitor | | Descriptor: | (1R,2S)-2-phenylcyclopropanamine, 2-methylsulfanylpyrimidine-4-carbaldehyde, Adenomatous polyposis coli protein, ... | | Authors: | Zhang, J, Wang, X.F, Song, K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | APC-Asef tripeptide inhibitor
To Be Published
|
|
8HTX
 
 | | Crystal structure of BANP in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*(5CM)P*GP*CP*GP*AP*GP*AP*G)-3'), Protein BANP | | Authors: | Zhang, J, Min, J, Liu, K. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
5H3V
 
 | | Crystal structure of a Type IV Secretion System Component CagX in Helicobacter pylori | | Descriptor: | Cag8, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL | | Authors: | Zhang, J, Wu, Y, Zhao, Y, Sun, L, Keegan, R.M, Liu, Y, Isupov, M.N. | | Deposit date: | 2016-10-27 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the type IV secretion system component CagX from Helicobacter pylori
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6LZ1
 
 | | Structure of S.pombe alpha-mannosidase Ams1 | | Descriptor: | Ams1, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of fission yeast tetrameric alpha-mannosidase Ams1.
Febs Open Bio, 10, 2020
|
|
7V9G
 
 | | Native BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*TP*GP*C)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9H
 
 | | The BEN3 domain of protein Bend3 | | Descriptor: | BEN domain-containing protein 3 | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9F
 
 | | Selenomethionine mutant (L740Sem) of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, CITRIC ACID, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), ... | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
5SVD
 
 | |
5T9P
 
 | |
3UIT
 
 | | Overall structure of Patj/Pals1/Mals complex | | Descriptor: | ACETATE ION, InaD-like protein, MAGUK p55 subfamily member 5, ... | | Authors: | Zhang, J, Yang, X, Long, J, Shen, Y. | | Deposit date: | 2011-11-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of an L27 domain heterotrimer from cell polarity complex Patj/Pals1/Mals2 reveals mutually independent L27 domain assembly mode
J.Biol.Chem., 287, 2012
|
|
