5ZV3
 
 | |
5XP9
 
 | | Crystal structure of Bismuth bound NDM-1 | | Descriptor: | Bismuth(III) ION, GLYCEROL, Metallo-beta-lactamase type 2 | | Authors: | Zhang, H, Ma, G, Lai, T.P, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
5XP6
 
 | | native structure of NDM-1 crystallized at pH5.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G, Lai, J, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
8KCB
 
 | | Complex of DDM1-nucleosome(H2A) complex with DDM1 bound to SHL2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Zhang, H, Zhang, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Mechanism of heterochromatin remodeling revealed by the DDM1 bound nucleosome structures.
Structure, 32, 2024
|
|
8KCC
 
 | | Complex of DDM1-nucleosome(H2A.W) complex with DDM1 bound to SHL2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Zhang, H, Zhang, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of heterochromatin remodeling revealed by the DDM1 bound nucleosome structures.
Structure, 32, 2024
|
|
5GS5
 
 | | Crystal structure of apo rat STING | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of apo ratSTING
To Be Published
|
|
5GRM
 
 | | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP) | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-11 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP)
To Be Published
|
|
5ZGU
 
 | |
5ZGW
 
 | |
5ZGY
 
 | |
5XAG
 
 | | Crystal structure of tubulin-stathmin-TTL-Compound Z2 complex | | Descriptor: | (3~{R},4~{R})-3-(hydroxymethyl)-4-(4-methoxy-3-oxidanyl-phenyl)-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
5ZGX
 
 | |
5ZGF
 
 | | Crystal structure of NDM-1 Q123G mutant | | Descriptor: | HYDROXIDE ION, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGI
 
 | |
5ZIA
 
 | |
5ZGT
 
 | |
5ZH1
 
 | |
5ZGQ
 
 | | Crystal structure of NDM-1 at pH7.5 (Tris-HCl, (NH4)2SO4) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGZ
 
 | |
5ZGP
 
 | | Crystal structure of NDM-1 at pH6.2 (Bis-Tris) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
5ZGR
 
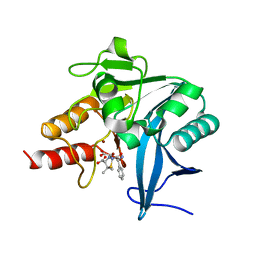 | | Crystal structure of NDM-1 at pH7.3 (HEPES) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGV
 
 | |
5XAF
 
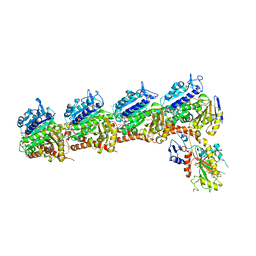 | | Crystal structure of tubulin-stathmin-TTL-Compound Z1 complex | | Descriptor: | (3S,4R)-4-(3-hydroxy-4-methoxyphenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
8FK2
 
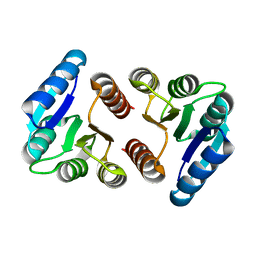 | | The N-terminal VicR from Streptococcus mutans | | Descriptor: | Putative response regulator CovR VicR-like protein | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Small Molecule Attenuates Bacterial Virulence by Targeting Conserved Response Regulator.
Mbio, 14, 2023
|
|
