6NMA
 
 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NM9
 
 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4 dimer | | Descriptor: | AcrVA4, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NIH
 
 | | Crystal structure of human TLR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Su, L, Zhang, H. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of TLR2/TLR1 Activation by the Synthetic Agonist Diprovocim.
J. Med. Chem., 62, 2019
|
|
6NME
 
 | | Structure of LbCas12a-crRNA | | Descriptor: | Cpf1, MAGNESIUM ION, crRNA | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.67 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
2I7X
 
 | |
5VJX
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
5VJI
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-07 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
6NMC
 
 | | CryoEM structure of the LbCas12a-crRNA-2xAcrVA1 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
8WU1
 
 | | Cryo-EM structure of CB1-beta-arrestin1 complex | | Descriptor: | Beta-arrestin-1, Cannabinoid receptor 1,Vasopressin V2 receptor, Fab30 heavy chain, ... | | Authors: | Liao, Y, Zhang, H, Shen, Q, Cai, C. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Snapshot of the cannabinoid receptor 1-arrestin complex unravels the biased signaling mechanism.
Cell, 186, 2023
|
|
8WZ2
 
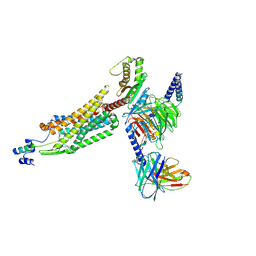 | | Structure of 26RFa-pyroglutamylated RFamide peptide receptor complex | | Descriptor: | G-alpha q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jin, S, Li, X, Xu, Y, Guo, S, Wu, C, Zhang, H, Yuan, Q, Xu, H.E, Xie, X, Jiang, Y. | | Deposit date: | 2023-11-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for recognition of 26RFa by the pyroglutamylated RFamide peptide receptor.
Cell Discov, 10, 2024
|
|
3FIU
 
 | | Structure of NMN synthetase from Francisella tularensis | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Sorci, L, Martynowski, D, Eyobo, Y, Osterman, A.L, Zhang, H. | | Deposit date: | 2008-12-12 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nicotinamide mononucleotide synthetase is the key enzyme for an alternative route of NAD biosynthesis in Francisella tularensis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
2HBX
 
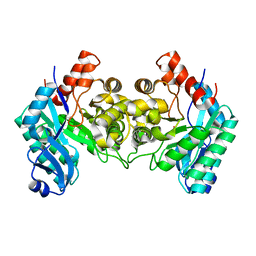 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
7WJQ
 
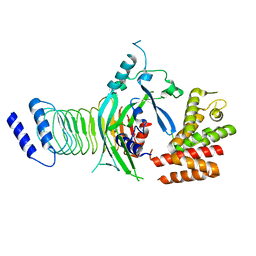 | | Crystal structure of GSDMB in complex with Ipah7.8 | | Descriptor: | Isoform 2 of Gasdermin-B, Probable E3 ubiquitin-protein ligase ipaH7.8 | | Authors: | Li, X, Zhang, H, Yin, H. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the GSDMB-mediated cellular lysis and its targeting by IpaH7.8
Nat Commun, 14, 2023
|
|
3G59
 
 | |
3G6K
 
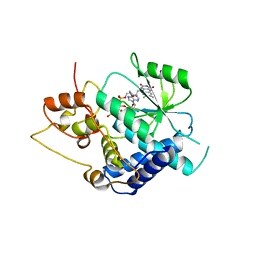 | | Crystal Structure of Candida glabrata FMN Adenylyltransferase in complex with FAD and Inorganic Pyrophosphate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FMN adenylyltransferase, MAGNESIUM ION, ... | | Authors: | Huerta, C, Machius, M, Zhang, H. | | Deposit date: | 2009-02-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and mechanism of a eukaryotic FMN adenylyltransferase.
J.Mol.Biol., 389, 2009
|
|
3FWK
 
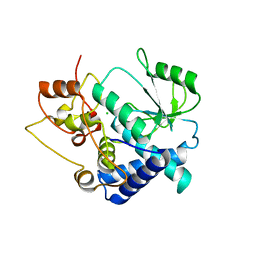 | |
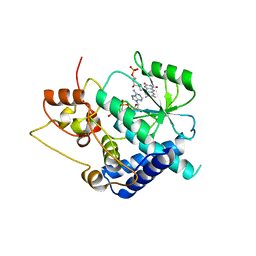
3G5A
 
 | |
8IA7
 
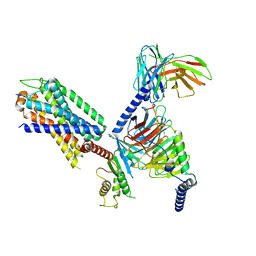 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | CCK-8, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7WDL
 
 | | Fungal immunomodulatory protein FIP-nha | | Descriptor: | Fungal immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties.
Int.J.Biol.Macromol., 213, 2022
|
|
5GRR
 
 | | Crystal structure of MCR-1 | | Descriptor: | GLYCEROL, Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Ma, G, Zhu, Y, Yu, Z, Zhang, H. | | Deposit date: | 2016-08-12 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High resolution crystal structure of the catalytic domain of MCR-1
Sci Rep, 6, 2016
|
|
5GJJ
 
 | |
5GYY
 
 | | Plant receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S-locus protein 11, S-receptor kinase SRK9 | | Authors: | Ma, R, Han, Z, Hu, Z, Lin, G, Gong, X, Zhang, H, June, N, Chai, J. | | Deposit date: | 2016-09-24 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Plant receptor complex at 2.35 Angstroms resolution
To Be Published
|
|
5GNI
 
 | |
5H49
 
 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
