5H49
 
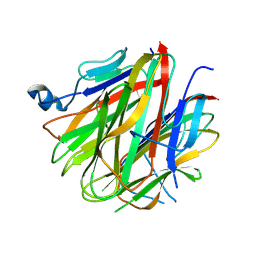 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5IC0
 
 | | Structural analysis of a talin triple domain module | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Brennan, M.L. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
4F3L
 
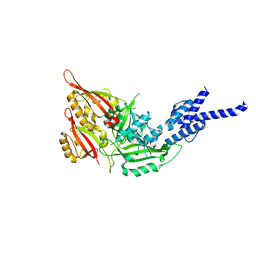 | | Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex | | Descriptor: | BMAL1b, Circadian locomoter output cycles protein kaput | | Authors: | Huang, N, Chelliah, Y, Shan, Y, Taylor, C, Yoo, S, Partch, C, Green, C.B, Zhang, H, Takahashi, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex.
Science, 337, 2012
|
|
5I8J
 
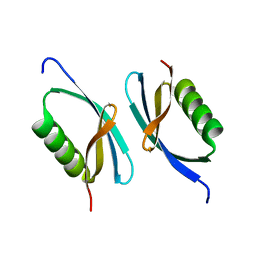 | | Crystal structure of Dmd from phage RB69 | | Descriptor: | Dmd discriminator of mRNA degradation | | Authors: | Wei, Y, Zhang, H, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterizations of phage antitoxin Dmd and its interactions with bacterial toxin RnlA
Biochem. Biophys. Res. Commun., 472, 2016
|
|
4HE9
 
 | | Crystal Structure of HIV-1 protease mutants I54M complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 protease, ... | | Authors: | Shen, C.H, Zhang, H, Weber, I.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
5E6N
 
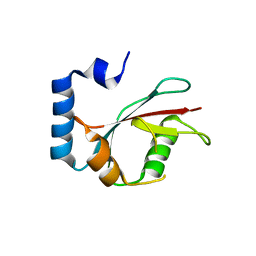 | | Crystal structure of C. elegans LGG-2 | | Descriptor: | Protein lgg-2 | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
5IC1
 
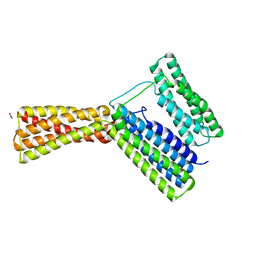 | | Structural analysis of a talin triple domain module, E1794Y, E1797Y, Q1801Y mutant | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Huang, Q.-Q. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
5HNT
 
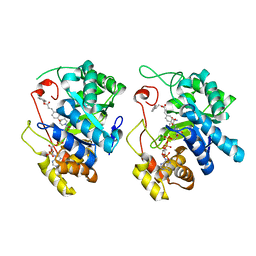 | | Crystal Structure of AKR1C3 complexed with CAPE | | Descriptor: | 2-phenylethyl (2E)-3-(3,4-dihydroxyphenyl)prop-2-enoate, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C, Zhao, Y, Zhang, H, Hu, X. | | Deposit date: | 2016-01-18 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of AKR1C3 complexed with octyl gallate
To Be Published
|
|
5HNU
 
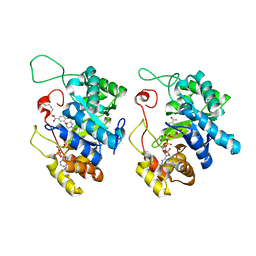 | | Crystal Structure of AKR1C3 complexed with octyl gallate | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, octyl 3,4,5-trihydroxybenzoate | | Authors: | Li, C, Zhao, Y, Zhang, H, Hu, X. | | Deposit date: | 2016-01-18 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of AKR1C3 complexed with octyl gallte
To Be Published
|
|
5GRR
 
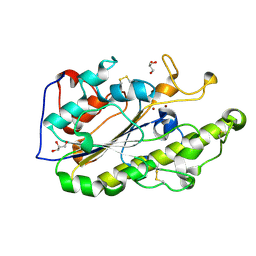 | | Crystal structure of MCR-1 | | Descriptor: | GLYCEROL, Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Ma, G, Zhu, Y, Yu, Z, Zhang, H. | | Deposit date: | 2016-08-12 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High resolution crystal structure of the catalytic domain of MCR-1
Sci Rep, 6, 2016
|
|
5H48
 
 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H4C
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Cbln4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
4HDP
 
 | | Crystal Structure of HIV-1 protease mutants I50V complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Shen, C.H, Zhang, H, Weber, I.T. | | Deposit date: | 2012-10-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4HZ9
 
 | | Crystal structure of the type VI native effector-immunity complex Tae3-Tai3 from Ralstonia pickettii | | Descriptor: | Putative cytoplasmic protein, Putative periplasmic protein | | Authors: | Dong, C, Zhang, H, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2012-11-14 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the inhibition of type VI effector Tae3 by its immunity protein Tai3
Biochem.J., 454, 2013
|
|
6LG4
 
 | |
6LG8
 
 | |
3OJW
 
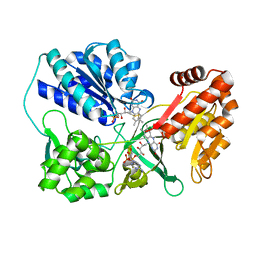 | | Disulfide crosslinked cytochrome P450 reductase inactive | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-cytochrome P450 reductase | | Authors: | Xia, C, Hamdane, D, Shen, A, Choi, V, Kasper, C, Zhang, H, Im, S.-C, Waskell, L, Kim, J.-J.P. | | Deposit date: | 2010-08-23 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes of NADPH-Cytochrome P450 Oxidoreductase Are Essential for Catalysis and Cofactor Binding.
J.Biol.Chem., 286, 2011
|
|
3Q6B
 
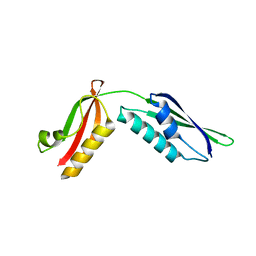 | |
5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
3QB5
 
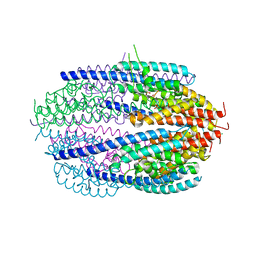 | | Human C3PO complex in the presence of MnSO4 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Ye, X, Huang, N, Liu, Y, Paroo, Z, Chen, S, Zhang, H, Liu, Q. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of C3PO and mechanism of human RISC activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3Q5Y
 
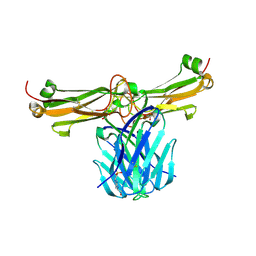 | | V beta/V beta homodimerization-based pre-TCR model suggested by TCR beta crystal structures | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Q, Zhang, H, Wang, J.-H. | | Deposit date: | 2010-12-30 | | Release date: | 2011-03-09 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A conserved hydrophobic patch on Vbeta domains revealed by TCRbeta chain crystal structures: implications for pre-TCR dimerization
Front Immunol, 2, 2011
|
|
3OJX
 
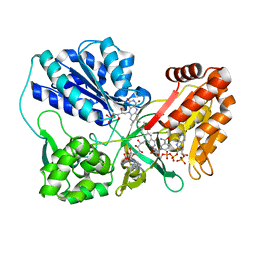 | | Disulfide crosslinked cytochrome P450 reductase inactive | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Hamdane, D, Shen, A, Choi, V, Kasper, C, Zhang, H, Im, S.-C, Waskell, L, Kim, J.-J.P. | | Deposit date: | 2010-08-23 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes of NADPH-Cytochrome P450 Oxidoreductase Are Essential for Catalysis and Cofactor Binding.
J.Biol.Chem., 286, 2011
|
|
3Q5T
 
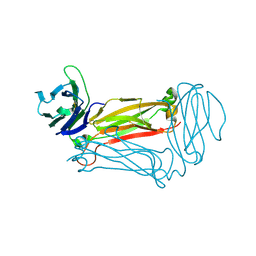 | |
3PJA
 
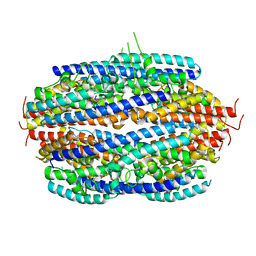 | | Crystal structure of human C3PO complex | | Descriptor: | Translin, Translin-associated protein X | | Authors: | Huang, N, Zhang, H. | | Deposit date: | 2010-11-09 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of C3PO and mechanism of human RISC activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4O5P
 
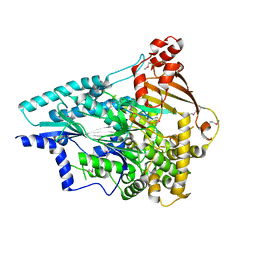 | | Crystal structure of an uncharacterized protein from Pseudomonas aeruginosa | | Descriptor: | Uncharacterized protein | | Authors: | Hu, H.D, Gao, Z.Q, Zhang, H, Dong, Y.H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-08-13 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structure of the type VI secretion phospholipase effector Tle1 provides insight into its hydrolysis and membrane targeting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
