4OV1
 
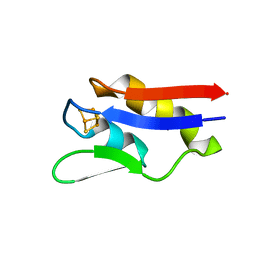 | | The crystal structure of a novel electron transfer ferredoxin from R. palustris HaA2 | | 分子名称: | FE3-S4 CLUSTER, Putative ferredoxin | | 著者 | Zhouw, W.H, Zhang, T, Zhang, A.L, Bell, S.G, Wong, L.-L. | | 登録日 | 2014-02-19 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.306 Å) | | 主引用文献 | The structure of a novel electron-transfer ferredoxin from Rhodopseudomonas palustris HaA2 which contains a histidine residue in its iron-sulfur cluster-binding motif.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5KZN
 
 | | Metabotropic Glutamate Receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 2 | | 著者 | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | 登録日 | 2016-07-25 | | 公開日 | 2016-12-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
5KZQ
 
 | | Metabotropic Glutamate Receptor in complex with antagonist (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid | | 分子名称: | (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | 著者 | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | 登録日 | 2016-07-25 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
4DO1
 
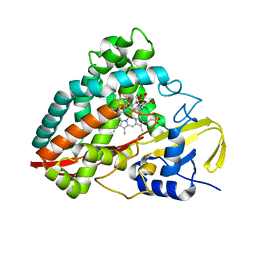 | | The crystal structures of 4-methoxybenzoate bound CYP199A4 | | 分子名称: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | 著者 | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | 登録日 | 2012-02-09 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4DNJ
 
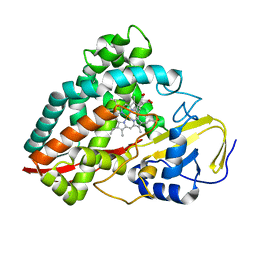 | | The crystal structures of 4-methoxybenzoate bound CYP199A2 | | 分子名称: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | 登録日 | 2012-02-08 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4I5I
 
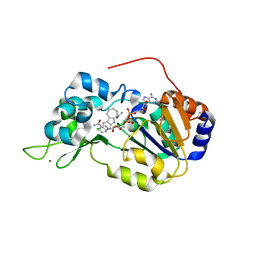 | | Crystal structure of the SIRT1 catalytic domain bound to NAD and an EX527 analog | | 分子名称: | (6S)-2-chloro-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Zhao, X, Allison, D, Condon, B, Zhang, F, Gheyi, T, Zhang, A, Ashok, S, Russell, M, Macewan, I, Qian, Y, Jamison, J.A, Luz, J.G. | | 登録日 | 2012-11-28 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The 2.5 angstrom crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition.
J.Med.Chem., 56, 2013
|
|
6BSZ
 
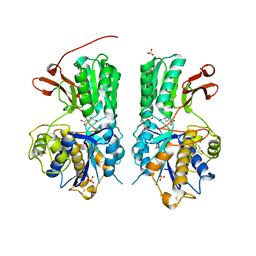 | | Human mGlu8 Receptor complexed with glutamate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 8, ... | | 著者 | Schkeryantz, J.M, Chen, Q, Ho, J.D, Atwell, S, Zhang, A, Vargas, M.C, Wang, J, Monn, J.A, Hao, J. | | 登録日 | 2017-12-04 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Determination of L-AP4-bound human mGlu8 receptor amino terminal domain structure and the molecular basis for L-AP4's group III mGlu receptor functional potency and selectivity.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6BT5
 
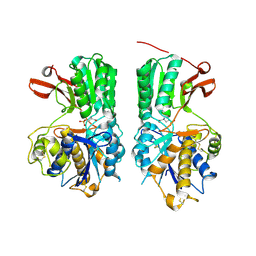 | | Human mGlu8 Receptor complexed with L-AP4 | | 分子名称: | (2S)-2-amino-4-phosphonobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 8 | | 著者 | Schkeryantz, J.M, Chen, Q, Ho, J.D, Atwell, S, Zhang, A, Vargas, M.C, Wang, J, Monn, J.A, Hao, J. | | 登録日 | 2017-12-05 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Determination of L-AP4-bound human mGlu8 receptor amino terminal domain structure and the molecular basis for L-AP4's group III mGlu receptor functional potency and selectivity.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
3C6F
 
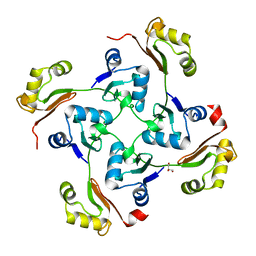 | | Crystal structure of protein Bsu07140 from Bacillus subtilis | | 分子名称: | GLYCEROL, YetF protein | | 著者 | Patskovsky, Y, Min, T, Zhang, A, Adams, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-02-04 | | 公開日 | 2008-02-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of protein Bsu07140 from Bacillus subtilis.
To be Published
|
|
6VGI
 
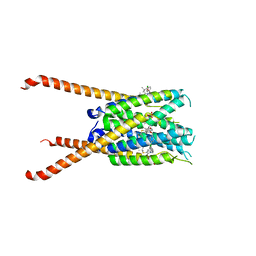 | | Crystal Structures of FLAP bound to MK-866 | | 分子名称: | 3-[3-(tert-butylsulfanyl)-1-[(4-chlorophenyl)methyl]-5-(propan-2-yl)-1H-indol-2-yl]-2,2-dimethylpropanoic acid, 5-lipoxygenase-activating protein, SULFATE ION | | 著者 | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | 登録日 | 2020-01-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VGC
 
 | | Crystal Structures of FLAP bound to DG-031 | | 分子名称: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, 5-lipoxygenase-activating protein, CALCIUM ION, ... | | 著者 | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | 登録日 | 2020-01-07 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VL4
 
 | | Crystal Structure of mPGES-1 bound to DG-031 | | 分子名称: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, Prostaglandin E synthase, TETRAETHYLENE GLYCOL, ... | | 著者 | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | 登録日 | 2020-01-22 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
3E63
 
 | | Fragment based discovery of JAK-2 inhibitors | | 分子名称: | 5-phenyl-1H-indazol-3-amine, Tyrosine-protein kinase JAK2 | | 著者 | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | 登録日 | 2008-08-14 | | 公開日 | 2008-10-14 | | 最終更新日 | 2012-02-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E62
 
 | | Fragment based discovery of JAK-2 inhibitors | | 分子名称: | 5-bromo-1H-indazol-3-amine, Tyrosine-protein kinase JAK2 | | 著者 | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | 登録日 | 2008-08-14 | | 公開日 | 2008-10-14 | | 最終更新日 | 2012-02-08 | | 実験手法 | X-RAY DIFFRACTION (1.922 Å) | | 主引用文献 | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3I3G
 
 | | Crystal Structure of Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A | | 分子名称: | N-acetyltransferase | | 著者 | Qiu, W, Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, J Ferguson, M.A, Hui, R, Structural Genomics Consortium (SGC) | | 登録日 | 2009-06-30 | | 公開日 | 2009-08-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal Structure Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A
To be Published
|
|
3E64
 
 | | Fragment based discovery of JAK-2 inhibitors | | 分子名称: | 4-(3-amino-1H-indazol-5-yl)-N-tert-butylbenzenesulfonamide, Tyrosine-protein kinase JAK2 | | 著者 | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | 登録日 | 2008-08-14 | | 公開日 | 2008-10-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4GQB
 
 | | Crystal Structure of the human PRMT5:MEP50 Complex | | 分子名称: | (2S,5S,6E)-2,5-diamino-6-[(3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxydihydrofuran-2(3H)-ylidene]hexanoic acid, Histone H4 peptide, Methylosome protein 50, ... | | 著者 | Antonysamy, S, Bonday, Z, Campbell, R, Doyle, B, Druzina, Z, Gheyi, T, Han, B, Jungheim, L.N, Qian, Y, Rauch, C, Russell, M, Sauder, J.M, Wasserman, S.R, Weichert, K, Willard, F.S, Zhang, A, Emtage, S. | | 登録日 | 2012-08-22 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Crystal structure of the human PRMT5:MEP50 complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6KGS
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.309 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
5TUD
 
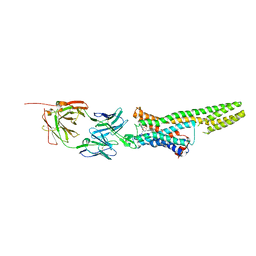 | | Structural Insights into the Extracellular Recognition of the Human Serotonin 2B Receptor by an Antibody | | 分子名称: | 5-hydroxytryptamine receptor 2B,Soluble cytochrome b562 chimera, Anti-5-HT2B Fab heavy chain, Anti-5-HT2B Fab light chain, ... | | 著者 | Ishchenko, A, Wacker, D, Kapoor, M, Zhang, A, Han, G.W, Basu, S, Boutet, S, James, D, Wang, D, Weierstall, U, Liu, W, Katritch, V, Stevens, R.C, Cherezov, V. | | 登録日 | 2016-11-05 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural insights into the extracellular recognition of the human serotonin 2B receptor by an antibody.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ULC
 
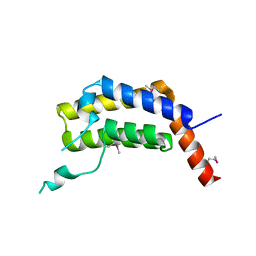 | | PLASMODIUM FALCIPARUM BROMODOMAIN-CONTAINING PROTEIN PF10_0328 | | 分子名称: | Bromodomain protein 1 | | 著者 | Wernimont, A.K, Amaya, M.F, Lam, A, Ali, A, Zhang, A.Z, Kenzina, L, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Weigelt, J, Hui, R, Walker, J.R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | 登録日 | 2017-01-24 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | PLASMODIUM FALCIPARUM BROMODOMAIN-CONTAINING PROTEIN PF10_0328
To be published
|
|
4DNZ
 
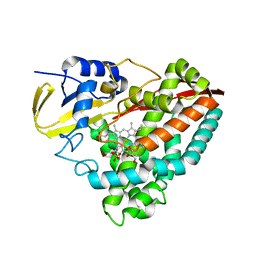 | | The crystal structures of CYP199A4 | | 分子名称: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | 著者 | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | 登録日 | 2012-02-09 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
6KGU
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with aztreonam | | 分子名称: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.106 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGV
 
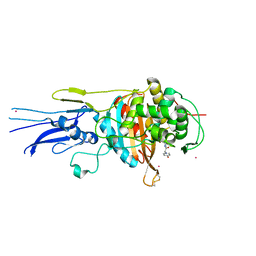 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with amoxicillin | | 分子名称: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, COBALT (II) ION, Penicillin-binding protein PbpB | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGH
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis (apo-form) | | 分子名称: | COBALT (II) ION, Penicillin-binding protein PbpB, SODIUM ION | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-11 | | 公開日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (2.108 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGW
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with ampicillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.407 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
