7VP7
 
 | |
7VP2
 
 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*TP*CP*CP*CP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*GP*GP*GP*AP*CP*CP*AP*CP*A)-3'), Transcription factor TCP10 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7VP4
 
 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*TP*CP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*GP*GP*GP*GP*AP*CP*CP*AP*CP*A)-3'), Transcription factor TCP10 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7VP6
 
 | |
7VP5
 
 | |
7VP3
 
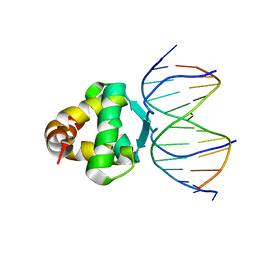 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*AP*CP*CP*CP*AP*C)-3'), Transcription factor TCP15 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7WFY
 
 | |
5UQ8
 
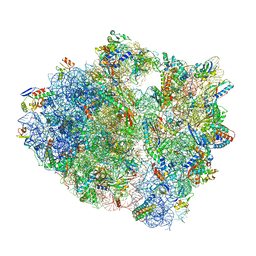 | | 70S ribosome complex with dnaX mRNA stem-loop and E-site tRNA ("out" conformation) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | Deposit date: | 2017-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2018-03-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
8HLC
 
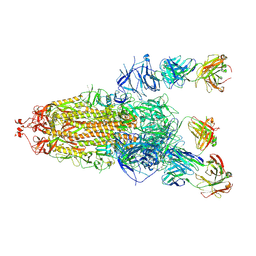 | | S protein of SARS-CoV-2 in complex with 3711 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A SARS-CoV-2 Spike N-terminal domain neutralizing antibody targets on a glycans shielded silent face and inhibits virus entry via hindering the recognition of RBD and hACE2
To Be Published
|
|
8HLD
 
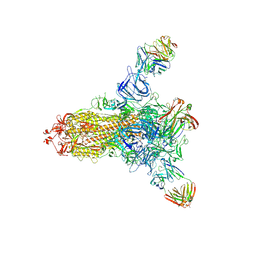 | | S protein of SARS-CoV-2 in complex with 26434 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A SARS-CoV-2 Spike N-terminal domain neutralizing antibody targets on a glycans shielded silent face and inhibits virus entry via hindering the recognition of RBD and hACE2
To Be Published
|
|
5UQ7
 
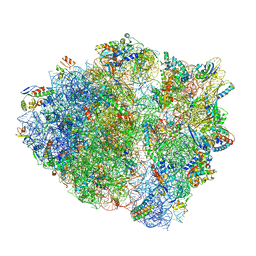 | | 70S ribosome complex with dnaX mRNA stemloop and E-site tRNA ("in" conformation) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | Deposit date: | 2017-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
7XRJ
 
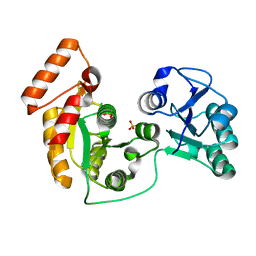 | | crystal structure of N-acetyltransferase DgcN-25328 | | Descriptor: | Putative NAD-dependent epimerase/dehydratase family protein, SULFATE ION | | Authors: | Zhang, Y.Z, Yu, Y, Cao, H.Y, Chen, X.L, Wang, P. | | Deposit date: | 2022-05-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel D-glutamate catabolic pathway in marine Proteobacteria and halophilic archaea.
Isme J, 17, 2023
|
|
7D0A
 
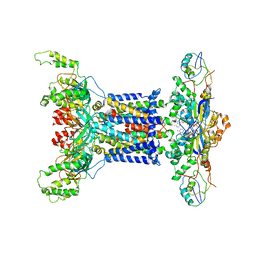 | | Acinetobacter MlaFEDB complex in ADP-vanadate trapped Vclose conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-DIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D09
 
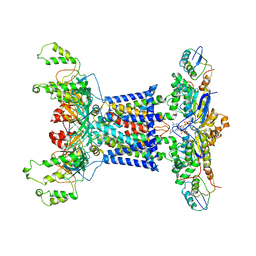 | | Acinetobacter MlaFEDB complex in ATP-bound Vtrans2 conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D08
 
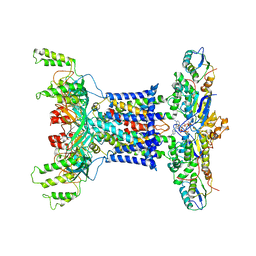 | | Acinetobacter MlaFEDB complex in ATP-bound Vtrans1 conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D06
 
 | | Cryo EM structure of the nucleotide free Acinetobacter MlaFEDB complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ABC transporter ATP-binding protein, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7C8G
 
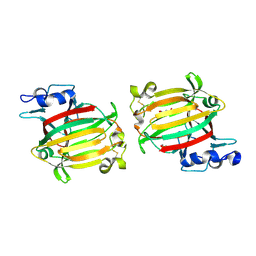 | | Structure of alginate lyase AlyC3 | | Descriptor: | Alginate lyase AlyC3, GLYCEROL, SUCCINIC ACID | | Authors: | Zhang, Y.Z, Xu, F, Chen, X.L, Wang, P. | | Deposit date: | 2020-05-30 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic.
J.Biol.Chem., 295, 2020
|
|
7XJR
 
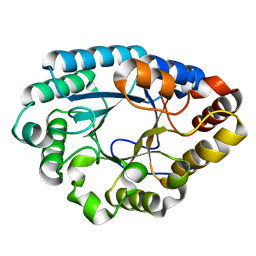 | | MLXase AlXyn26A | | Descriptor: | AlXyn26A | | Authors: | Zhang, Y.Z, Chen, X.L, Zhao, F, Yu, C.M. | | Deposit date: | 2022-04-18 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel class of xylanases specifically degrade marine red algal beta 1,3/1,4-mixed-linkage xylan.
J.Biol.Chem., 299, 2023
|
|
7XS3
 
 | | AlXyn26A E243A-X3X4X | | Descriptor: | AlXyn26A E243A-X3X4X, beta-D-xylopyranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Zhang, Y.Z, Chen, X.L, Zhao, F, Yu, C.M. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel class of xylanases specifically degrade marine red algal beta 1,3/1,4-mixed-linkage xylan.
J.Biol.Chem., 299, 2023
|
|
7F3J
 
 | |
7C8F
 
 | | Structure of alginate lyase AlyC3 in complex with dimannuronate(2M) | | Descriptor: | H127A/Y244A mutant of alginate lyase AlyC3 in complex with dimannuronate, MALONATE ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Zhang, Y.Z, Xu, F, Chen, X.L, Wang, P. | | Deposit date: | 2020-05-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic.
J.Biol.Chem., 295, 2020
|
|
7F3K
 
 | |
7F3I
 
 | |
7F3L
 
 | |
1Z33
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase | | Descriptor: | purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
