6IVR
 
 | | Crystal structure of a membrane protein W16A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
8UYO
 
 | |
7SGV
 
 | | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder630 inhibitor | | Descriptor: | CHLORIDE ION, N-(naphthalen-1-yl)pyridine-3-carboxamide, Papain-like protease, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder630 inhibitor
To Be Published
|
|
7SGU
 
 | | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder608 inhibitor | | Descriptor: | 5-amino-N-(naphthalen-1-yl)pyridine-3-carboxamide, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder608 inhibitor
To Be Published
|
|
7SGW
 
 | | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor | | Descriptor: | CHLORIDE ION, N-(naphthalen-1-yl)pyridine-3-carboxamide, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor
To Be Published
|
|
3JSG
 
 | |
8KCC
 
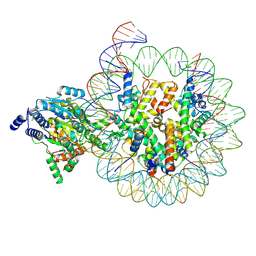 | | Complex of DDM1-nucleosome(H2A.W) complex with DDM1 bound to SHL2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Zhang, H, Zhang, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of heterochromatin remodeling revealed by the DDM1 bound nucleosome structures.
Structure, 32, 2024
|
|
6IVN
 
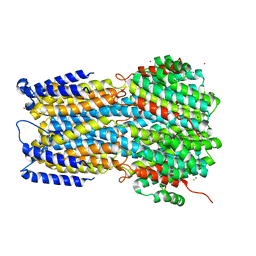 | | Crystal structure of a membrane protein G264A | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
7E3J
 
 | |
6IVL
 
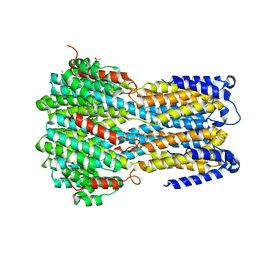 | | Crystal structure of a membrane protein L259A | | Descriptor: | ACETIC ACID, CHLORIDE ION, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
7XTB
 
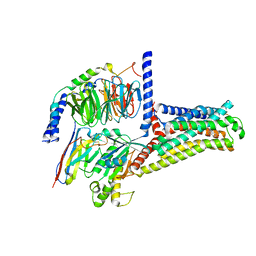 | | Serotonin 6 (5-HT6) receptor-Gs-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XTA
 
 | | Serotonin 4 (5-HT4) receptor-Gi-scFv16 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
6IVJ
 
 | | Crystal structure of a membrane protein G18A | | Descriptor: | ACETIC ACID, CHLORIDE ION, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
7XTC
 
 | | Serotonin 7 (5-HT7) receptor-Gs-Nb35 complex | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XT9
 
 | | Serotonin 4 (5-HT4) receptor-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XT8
 
 | | Serotonin 4 (5-HT4) receptor-Gs-Nb35 complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
4N6F
 
 | | Crystal structure of Amycolatopsis orientalis BexX complexed with G6P | | Descriptor: | CALCIUM ION, FRUCTOSE -6-PHOSPHATE, Putative thiosugar synthase | | Authors: | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-14 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
5V1X
 
 | | Carbon Sulfoxide lyase, Egt2 Y134F in complex with its substrate | | Descriptor: | (1S)-2-{2-[(R)-(2R)-2-amino-2-carboxyethanesulfinyl]-1H-imidazol-4-yl}-1-carboxy-N,N,N-trimethylethan-1-aminium, FORMIC ACID, Hercynylcysteine sulfoxide lyase | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.558 Å) | | Cite: | Snapshots of C-S Cleavage in Egt2 Reveals Substrate Specificity and Reaction Mechanism.
Cell Chem Biol, 25, 2018
|
|
8U2Y
 
 | | Solution structure of the PHD6 finger of MLL4 bound to TET3 | | Descriptor: | Histone-lysine N-methyltransferase 2D, Methylcytosine dioxygenase TET3, ZINC ION | | Authors: | Mohid, S.A, Zandian, M, Zhang, Y, Kutateladze, T.G. | | Deposit date: | 2023-09-06 | | Release date: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | MLL4 binds TET3.
Structure, 32, 2024
|
|
5YPS
 
 | | The structural basis of histone chaperoneVps75 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2017-11-03 | | Release date: | 2018-11-07 | | Last modified: | 2020-06-10 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
6VLS
 
 | | Structure of C-terminal fragment of Vip3A toxin | | Descriptor: | DI(HYDROXYETHYL)ETHER, Maltose/maltodextrin-binding periplasmic protein,Vip3Aa | | Authors: | Jiang, K, Zhang, Y, Chen, Z, Gao, X. | | Deposit date: | 2020-01-25 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Functional Insights into the C-terminal Fragment of Insecticidal Vip3A Toxin ofBacillus thuringiensis.
Toxins, 12, 2020
|
|
6LZE
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11a | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhang, Y, Jing, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-19 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
4Z4L
 
 | |
7N4Z
 
 | | Complex structure of NOS4 with noscapine | | Descriptor: | NOS4, SULFATE ION, noscapine | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
7N53
 
 | | Complex structure of PAP4 with papaverine | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, GLYCEROL, PAP4, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
